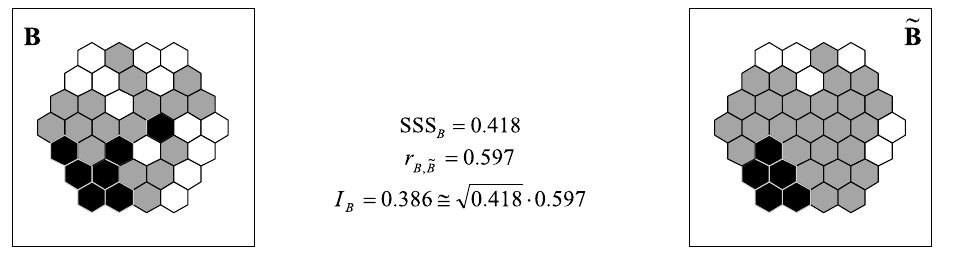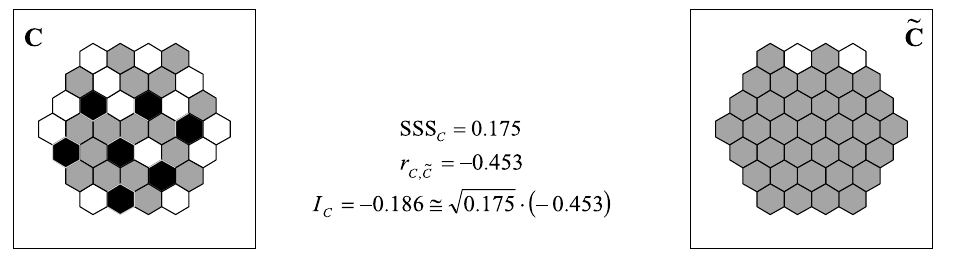Chapter 8 Bivariate relationships considering spatial pattern
Correlation coefficients are a well known measure of association between variables. The question is how these can or should be modified if we apply them to spatialy structured data. here we focus on association between numerical scaled variables and ignore association measures for ordinal or nominal variables.
8.1 Cheat sheet
The following packages are required for this chapter:
require(sf)
require(spdep)
require(sp)
require(ncf)
require(tmap)
require(ggplot2)
require(ggpubr)
require(gstat) # for simulation of autocorrelated data
require(dplyr)
require(SpatialPack) # for the modified t-testImportant commands:
spdep::lee.mc- permutation test for Lee’s Lspdep::lag.listw- computing a spatially lagged version of a variable vectorcor.test- Pearson’s r with test
8.3 Motivation
In presence of positive spatial autocorrelation our effective sample size is smaller than the sample size. This is due to shared information between observations. The normal t-test for Pearson’s r is therefore incorrect.
The simplest solution is to calculate the effective sample size under consideration of spatial autocorrelation.
The effective sample size \(n'\) in absence of autocorrelation: \(n' = n\) (number of observations). In presence of autocorrelation this needs to be modified to
\[n'= \frac{n^2}{\sum_{i=1}^n\sum_{j=1}^nCor(x_i, y_i)}\]
The exact formula depends on the from of the correlation function. For a first order autoregressive correlation structure (\(Cor(x_i, x_j)= \rho^{|i-j}\)):
\[n'=(1+2\frac{\rho}{1-\rho}(1-\frac{1}{n})-2\frac{\rho^2}{(1-\rho)^2} \frac{1-\rho^{n-1}}{n}) \cdot n\]
This can be simplified for large n:
\[n' \cong n \frac{1-\rho}{1+\rho}\]
# function to calculate the effective sampel size for large n and the simple first order autoregresive function
neff <- function(n, rho)
{
res <- n*(1-rho)/(1+rho)
return(res)
}- e.g. \(n=1000\) and \(\rho = 0.7\) \(\rightarrow n' =\) 176.4705882
- e.g. \(n=1000\) and \(\rho = 0.9\) \(\rightarrow n' =\) 52.6315789
The modified t-test by Clifford and Richardson (Clifford et al. (1989) and Dutilleul et al. (1993)) uses the covariance for all observations in a distance band as a measure to capture spatial autocorrelation (cf. correlogram). The function is available as spatialPack::modified.ttest.
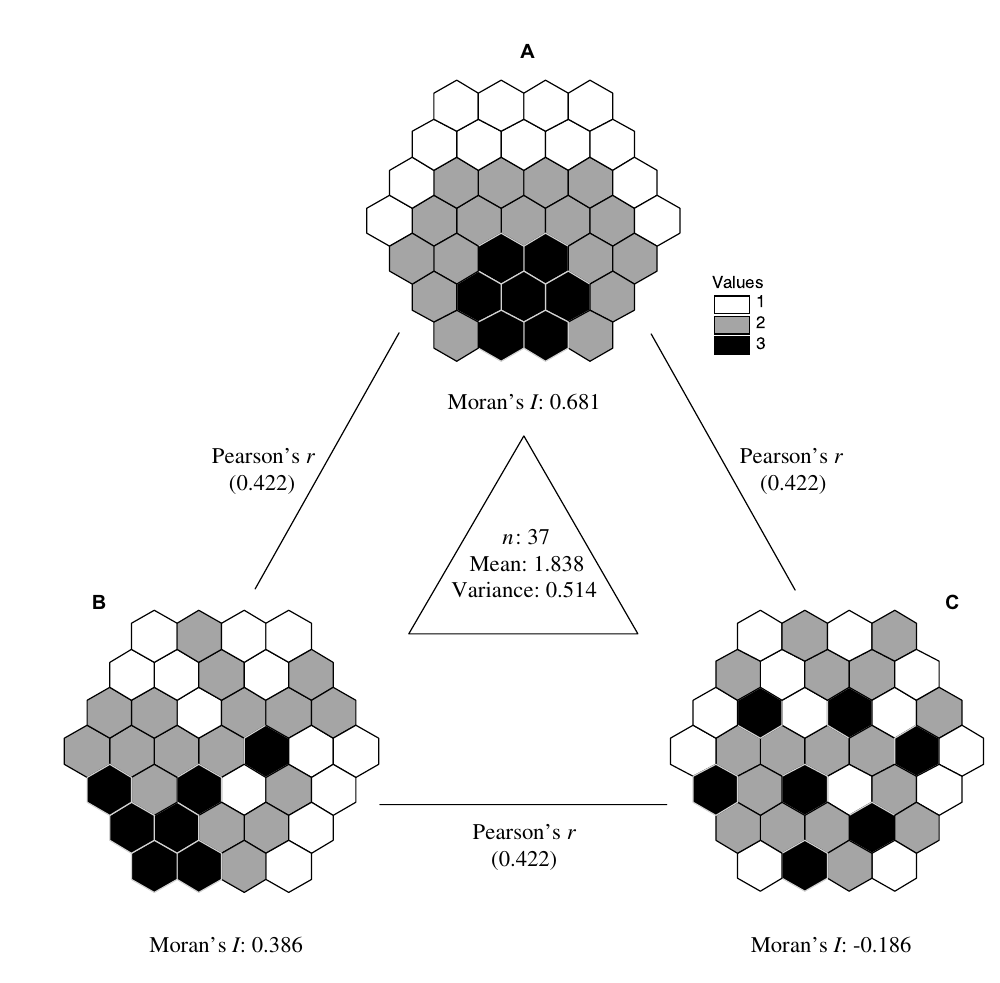
8.4 Bivariate Moran’s I
Bivariate Moran’s I is a measure of autocorrelation of X with the spatial lag of Y. As such, the resultant measure may overestimate the amount of spatial autocorrelation which may be a product of the inherent correlation of X and Y.
\[I_B = \frac{\sum_i(\sum_j w_{i,j}y_j\cdot x_i)}{\sum_i x_i^2}\] It only captures autocorrelation of one variable and completely ignores the spatial pattern of the second variable. \(I_{X,Y}\) might be different from \(I_{Y,X}\) which might provide an obstacle for interpretation. On the other hand, the bivariate Moran’s I looks at the relationship from a directed perspective - \(x\) depends on the lag of \(y\) - that might be interesting for spatial modelling.
Using the spatial lag operator this can be written as:
\[I_{X,Y} = \frac{\sum_i(x_i-\bar{x})(\tilde{y}-\bar{y})}{\sqrt{\sum_i(x_i-\bar{x})^2} \sqrt{\sum_i(y_i-\bar{y})^2}} \cong \sqrt{SSS_Y} \cdot r_{x,\tilde{y}}\]
8.6 Simulated data
We create a hexagon tesselation and add some spatially structure columns to it that can then be used to explore Lee’s L for these pattern.
set.seed(3)
aoi <- st_sfc(st_polygon(list(rbind(c(0,0), c(5,0), c(5,5), c(0,0)))))
hexgrid <- st_make_grid(aoi, cellsize = 1, square = FALSE)
hexgrid <- st_sf(hexgrid)
hexgrid <- st_sf(hexgrid, crs = 4326) # avoid warnings by assigning a CRS however, data is located nowhere
n <- nrow(hexgrid)
hexgrid$id <- 1:n
hexgrid <- hexgrid %>% mutate(var1 = ifelse(id <10, 30, ifelse(id < 20, 5, 1) + rnorm(n, mean= 0, sd=.1)),
var2 = ifelse(id <10, 2, ifelse(id < 20, 6, 20) + rnorm(n, mean= 0, sd=.1) ))
hexgrid <- hexgrid %>% mutate(var3 = rnorm(n, mean= 4) + var1,
var4 = rpois(n, lambda = var1),
var5 = rnorm(n, mean=30),
var6 = rnorm(n, mean=30))
hexgrid <- hexgrid %>% mutate(var7 = var6 + rnorm(n, mean= 0, sd=.5) )
idx <- sample(hexgrid$id, size=n, replace = FALSE)
hexgrid <- hexgrid %>% mutate(var8 = var1[idx], var9= var2[idx])
hexgrid <- hexgrid %>% mutate(var10 = var1 + rnorm(n))
hexgrid <- hexgrid %>% mutate(var11 = var10[idx])
hexgrid <- hexgrid %>% mutate(var1neg = max(var1)-1*var1)
hexgrid <- hexgrid %>% mutate(var12 = 10+ (id %%2) *10 - (id %%4)*2 - (id %%6)*2 + (id %% 8)*3)
hexgrid$var13 <- 20
hexgrid$var13[c(13, 14, 24, 35, 41, 30, 3, 5, 19, 40, 32, 18, 4)] <- 0
hexgrid$var13[c(23, 17,31, 37, 29, 15, 9, 17, 31, 43, 45 )] <- 40
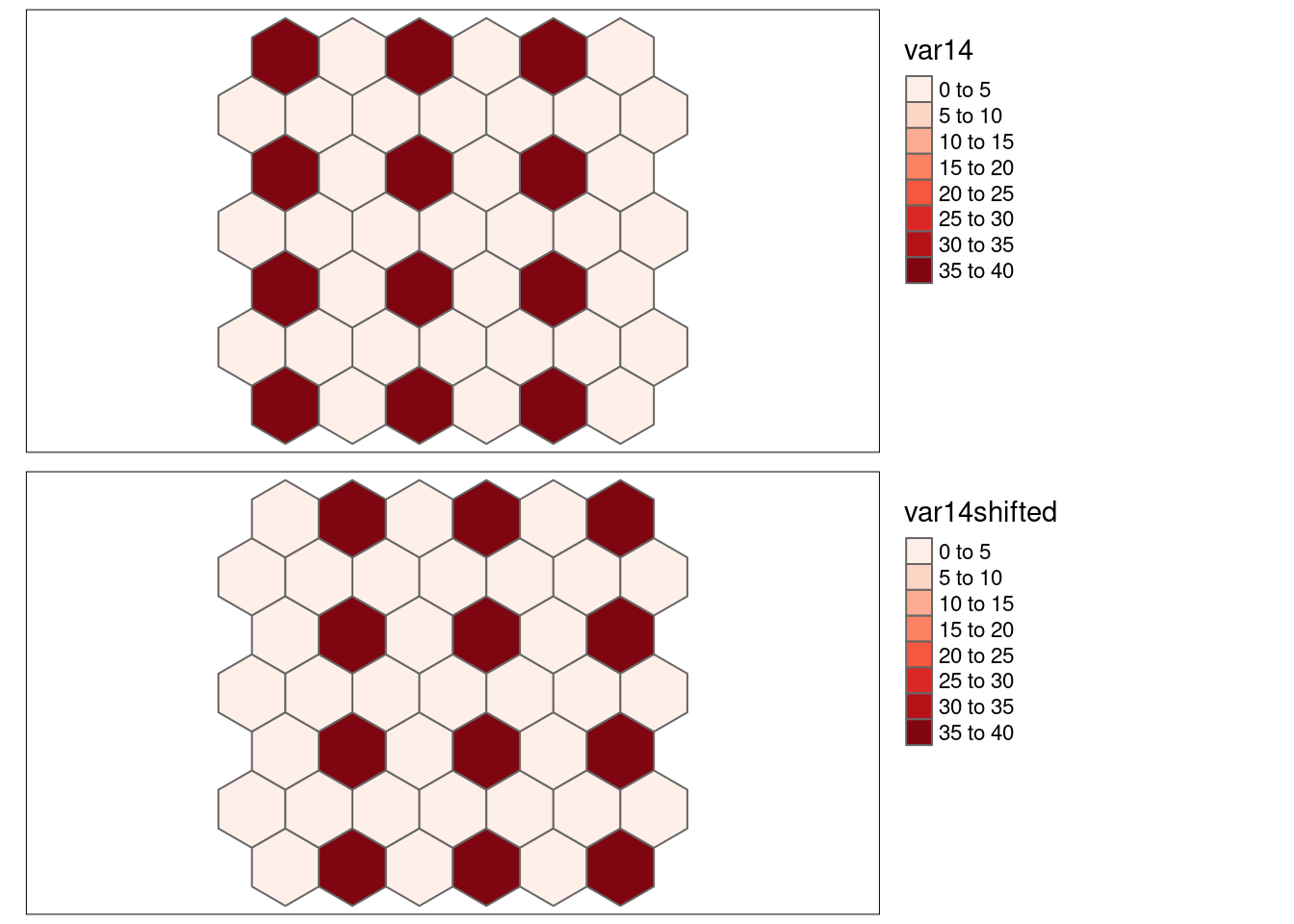
hexgrid$var14 <- 0
hexgrid$var14[c(4:7, 18:21, 32:35)] <- 40
#tm_shape(hexgrid) + tm_polygons(col="var14") + tm_layout(legend.outside = TRUE) + tm_text(text="id")
# shift values of var1 one hexcell to the right (+ 7 ids)
# see tm_shape(hexgrid) + tm_polygons(col="id") + tm_layout(legend.outside = TRUE) + tm_text(text="id")
moveHex1toLeft <- function(id)
{ # only valid for this little hexgrid
if(any(id>45)) {
stop("id needs to be smaller than 45")
}
newid <- ifelse( (id >=4) & (id <= 7),
id +35,
ifelse(id < 4,
id+42,
id-7))
return(newid)
}
hexgrid <- hexgrid %>% mutate(var1shifted = var1[moveHex1toLeft(id)],
var8shifted= var8[moveHex1toLeft(id)],
var13shifted= var13[moveHex1toLeft(id)],
var14shifted= var14[moveHex1toLeft(id)]
)The simulated data (cf. 8.2) contains strongly positively correlated autocorrelation (e.g. var1 and var2), negative autocorrelation (e.g. var12 and var13) and random pattern (var1-6). For some we have simply reshuffled the locations of the data to break spatial structure (var1 and var2 have been reshuffled to var8 and var9) while for others the pattern has been simply shifted by one cell to the right (the varXshifted variables) or the patterns has been flipped (var1neg).
theBreaks <-seq(0,40, by=5)
thePalette <- "brewer.reds"
m1 <- tm_shape(hexgrid) + tm_polygons(fill = "var1",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m2 <- tm_shape(hexgrid) + tm_polygons(fill="var2",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m3 <- tm_shape(hexgrid) + tm_polygons(fill="var3",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m4 <- tm_shape(hexgrid) + tm_polygons(fill="var4",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m5 <- tm_shape(hexgrid) + tm_polygons(fill="var5",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m6 <- tm_shape(hexgrid) + tm_polygons(fill="var6",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m7 <- tm_shape(hexgrid) + tm_polygons(fill="var7",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m8 <- tm_shape(hexgrid) + tm_polygons(fill="var8",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m9 <- tm_shape(hexgrid) + tm_polygons(fill="var9",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m10 <- tm_shape(hexgrid) + tm_polygons(fill="var10",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks )) +
tm_layout(legend.outside = TRUE)
m11 <- tm_shape(hexgrid) + tm_polygons(fill="var11",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m12 <- tm_shape(hexgrid) + tm_polygons(fill="var12",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m13 <- tm_shape(hexgrid) + tm_polygons(fill="var13",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m14 <- tm_shape(hexgrid) + tm_polygons(fill="var14",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m1neg <- tm_shape(hexgrid) + tm_polygons(fill="var1neg",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m1shifted <- tm_shape(hexgrid) + tm_polygons(fill="var1shifted",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m8shifted <- tm_shape(hexgrid) + tm_polygons(fill="var8shifted",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m13shifted <- tm_shape(hexgrid) + tm_polygons(fill="var13shifted",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
m14shifted <- tm_shape(hexgrid) + tm_polygons(fill="var14shifted",
fill.scale = tm_scale_intervals(values= thePalette,
breaks = theBreaks ),
fill.legend = tm_legend(reverse= TRUE)) +
tm_layout(legend.outside = TRUE)
tmap_arrange(m1,m2,m3,m4, m5, m6,m7, m8, m9, m10, m11, m12, m13, m14, m1neg, m1shifted, m8shifted, m13shifted, m14shifted)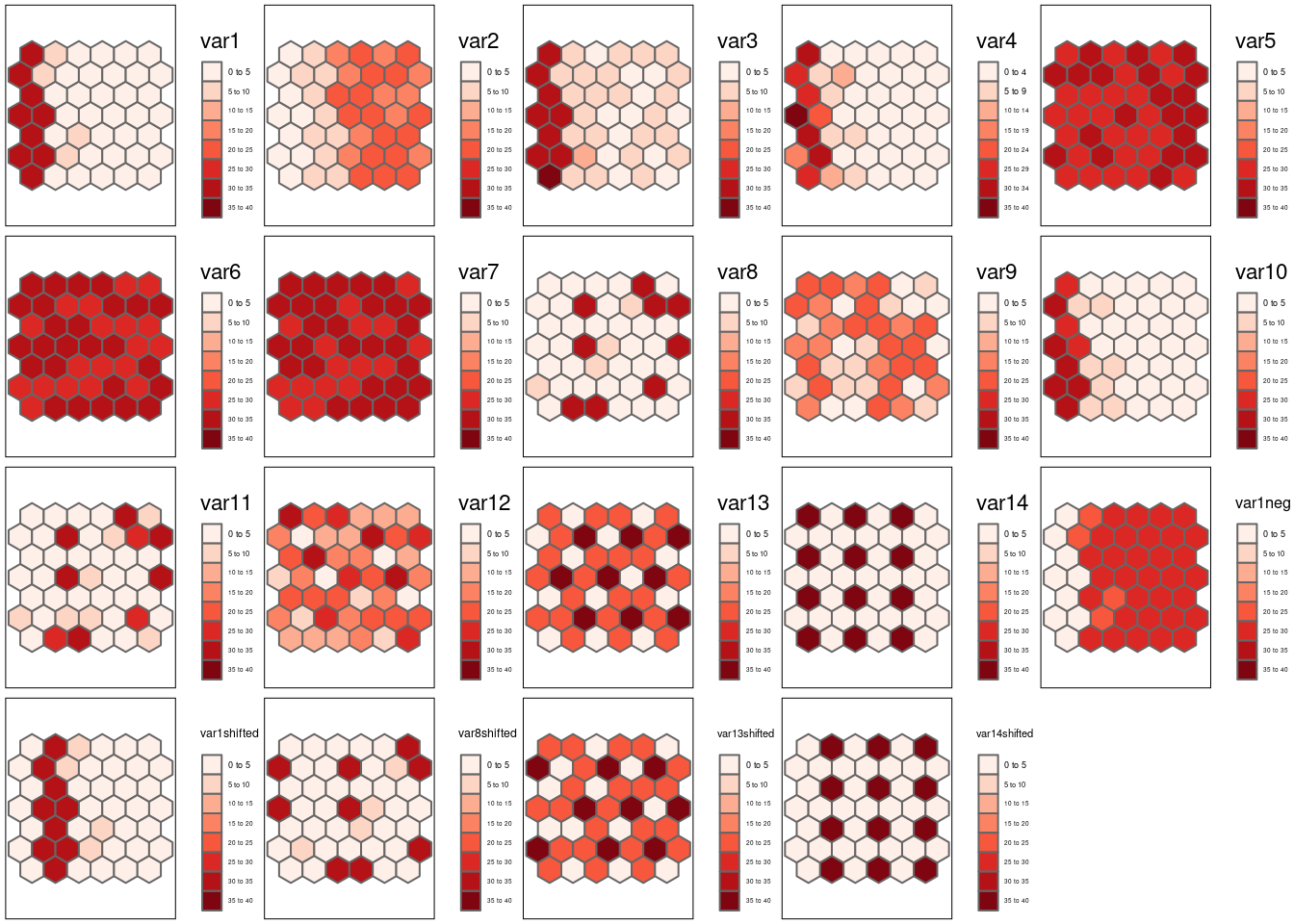
Figure 8.2: The following simulated patterns are analyzed later on with respect to Lee’s L.
A contiguity neighborhood definition is best suited for the case of the demonstration. This imples that each non-border cell has 6 neighbors while the border cells have less neighbors.
hexnb <- poly2nb(hexgrid)
hexlistw <- nb2listw(hexnb, style = "W")
hex_coord <- st_coordinates(hexgrid)[,1:2] # used later for the modified t-testIf we apply Moran’s test to the five simulated variables we see that all but the last three show significant spatial autocorrelation. (I am using sapply to map a function to a list but this is not of concern here.)
moran4vars <- lapply(2:8, FUN = function(x) moran.test(st_drop_geometry(hexgrid)[,x], listw = hexlistw))
moran4varsShorted <- sapply(moran4vars, FUN = function(x) c(x$estimate[1], x$p.value))
row.names(moran4varsShorted)[2] <- "p-value"
moran4varsShorted## [,1] [,2] [,3] [,4]
## Moran I statistic 6.778588e-01 8.140493e-01 6.773210e-01 6.101243e-01
## p-value 1.351183e-14 3.210743e-19 1.429188e-14 2.032328e-12
## [,5] [,6] [,7]
## Moran I statistic -0.007337836 0.01197456 0.1382407
## p-value 0.433931191 0.35233507 0.0370708Let us explore the case of the obviously autocorrelated and shifted first two variables:
##
## Monte-Carlo simulation of Lee's L
##
## data: hexgrid$var1 , hexgrid$var2
## weights: hexlistw
## number of simulations + 1: 500
##
## statistic = -0.63469, observed rank = 1, p-value = 0.002
## alternative hypothesis: less##
## Pearson's product-moment correlation
##
## data: hexgrid$var1 and hexgrid$var2
## t = -8.7979, df = 43, p-value = 3.624e-11
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## -0.8866491 -0.6646941
## sample estimates:
## cor
## -0.8017904Compared to Pearson’s r that ignores spatial autocorrelation Lee’s L indicates a weaker but still clear negative association between var1 and var2.
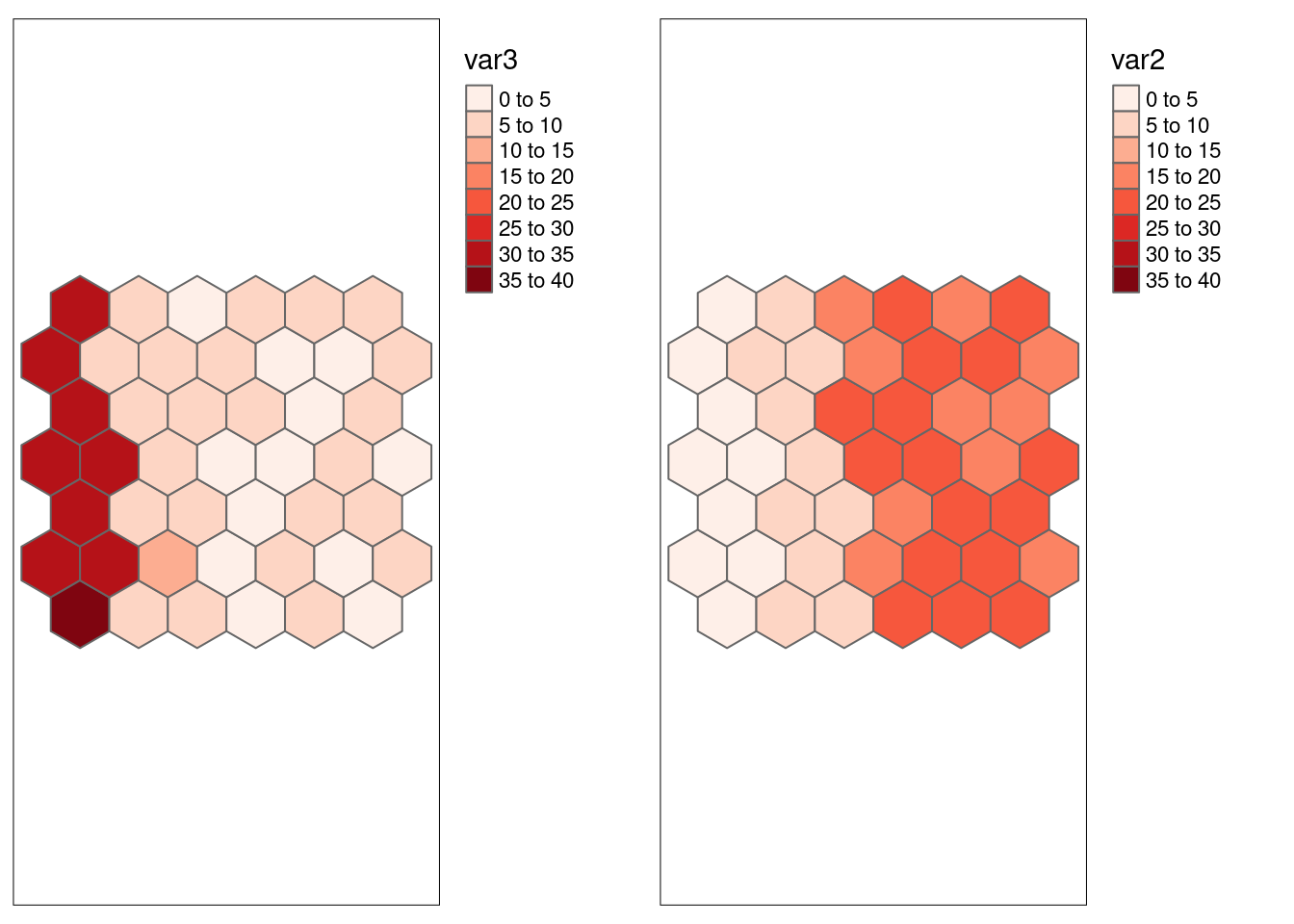
Let’s repeat that with var3 which has added white noise.
##
## Monte-Carlo simulation of Lee's L
##
## data: hexgrid$var3 , hexgrid$var2
## weights: hexlistw
## number of simulations + 1: 500
##
## statistic = -0.6389, observed rank = 1, p-value = 0.002
## alternative hypothesis: less##
## Pearson's product-moment correlation
##
## data: hexgrid$var3 and hexgrid$var2
## t = -8.828, df = 43, p-value = 3.294e-11
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## -0.8872331 -0.6662194
## sample estimates:
## cor
## -0.802766Similar. Both methods indicate significant negative correlation between var2 and var3. Lee’s L is against smaller due to the effect of positive spatial autocorrelation in the data.
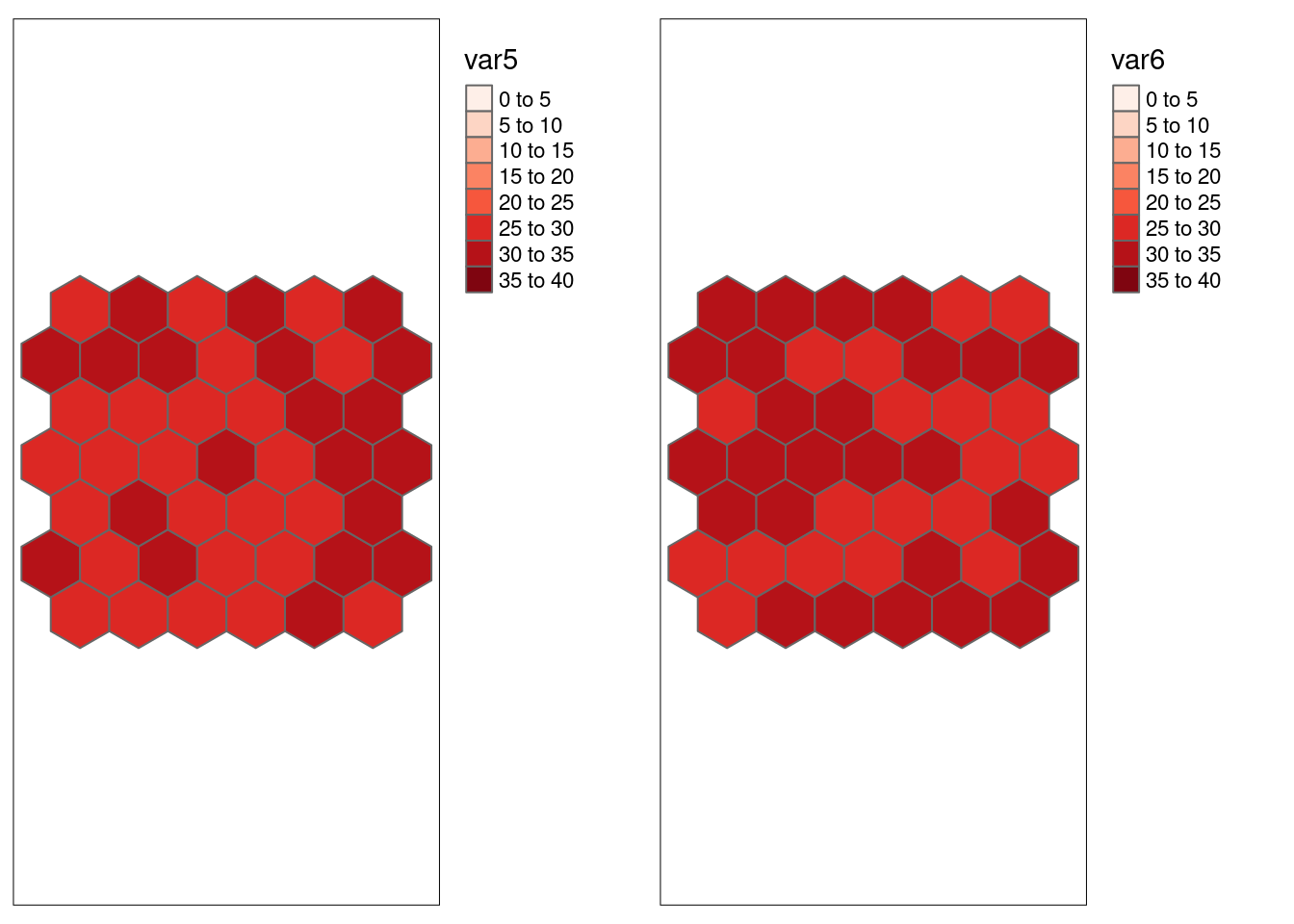
For the two white noise variables without spatial structure both association measures indicate expectedly no significant relationship.
##
## Monte-Carlo simulation of Lee's L
##
## data: hexgrid$var6 , hexgrid$var5
## weights: hexlistw
## number of simulations + 1: 500
##
## statistic = 0.061969, observed rank = 425, p-value = 0.85
## alternative hypothesis: less##
## Pearson's product-moment correlation
##
## data: hexgrid$var6 and hexgrid$var5
## t = 0.79209, df = 43, p-value = 0.4327
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## -0.179947 0.399396
## sample estimates:
## cor
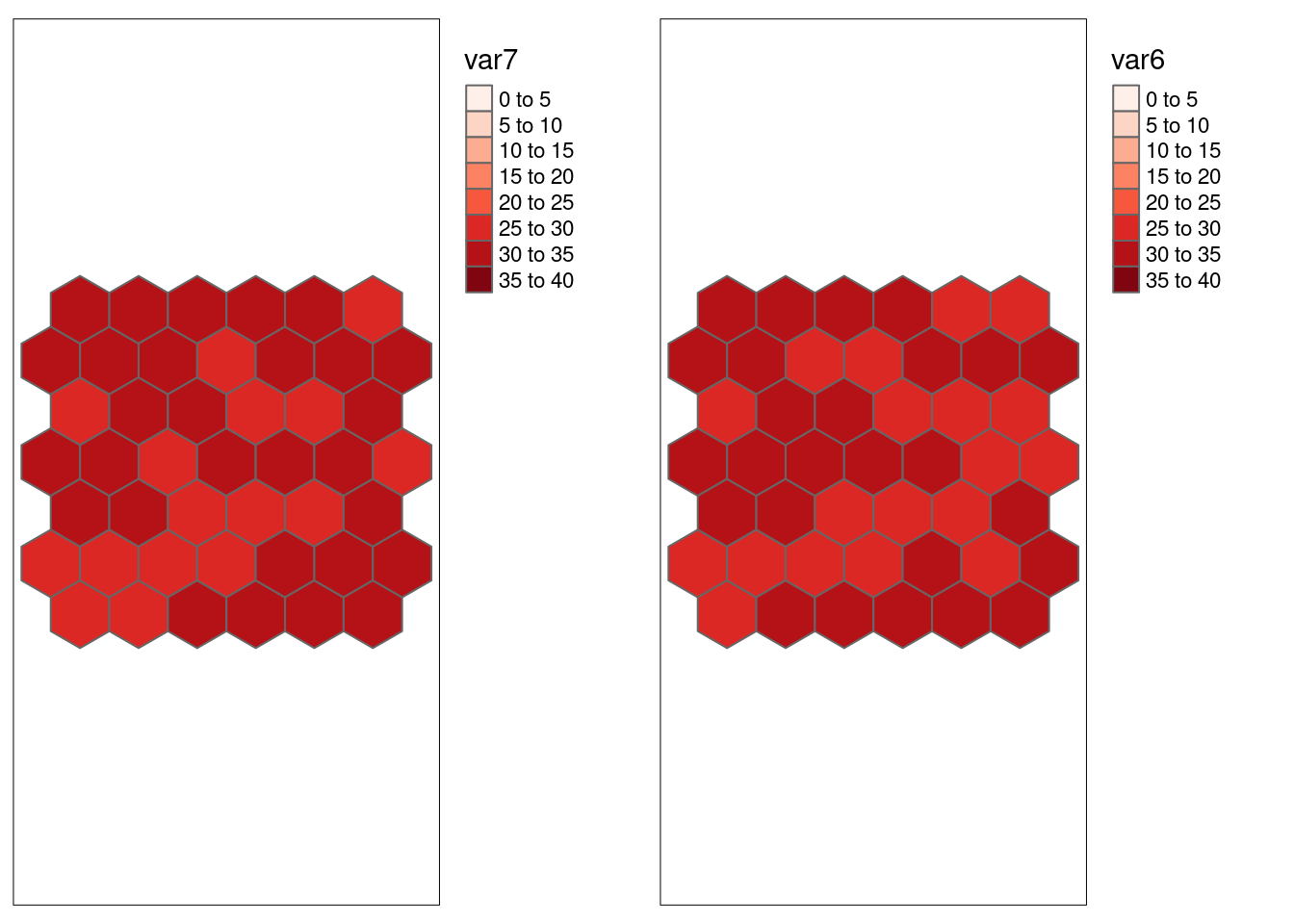
## 0.1199212The last case is interesting as we are comparing two variables without significant spatial autocorrelation that were designed to be correlated. While Pearson’s r indicates strong autocorrelation, Lee’´s L suggest no significant association.
##
## Monte-Carlo simulation of Lee's L
##
## data: hexgrid$var6 , hexgrid$var7
## weights: hexlistw
## number of simulations + 1: 1000
##
## statistic = 0.169, observed rank = 574, p-value = 0.426
## alternative hypothesis: greater##
## Pearson's product-moment correlation
##
## data: hexgrid$var6 and hexgrid$var7
## t = 8.9585, df = 43, p-value = 2.178e-11
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## 0.6727348 0.8897199
## sample estimates:
## cor
## 0.80692498.6.1 Examples
We are looking here at a number of cases in a bit more detail. The idea is to present simple cases that make it easier to spot and understand differences in the metrics calculated for the pattern. Of course, real world cases will be more complicated - if you think of those as a mixture of such archetypes of relationships it might help you to understand the meaning of Lee’s L.
This section is still a stub with very limited explanation. This should hopefully change in the not so far future.
8.6.1.1 Helper functions
To ease the code we define some functions for the calculation of the different indicators.
# functions for calculating spatial smoothing scalar
calcSSS <- function(values, listw, zero.policy=NULL)
{
# calc spatial lag
values.lag <- lag.listw(x=listw, var=values, zero.policy = zero.policy )
values.mean <- mean(values)
sssx <- sum((values.lag-values.mean)^2) / sum((values-values.mean)^2)
return(sssx)
}
calcPearsonLagged <- function(x,y, listw, zero.policy=NULL)
{
xlag<- lag.listw(x=listw, var=x, zero.policy = zero.policy )
ylag<- lag.listw(x=listw, var=y, zero.policy = zero.policy )
res <- cor(xlag, ylag)
return(res)
}
calcSpatialCorrIndicators <- function(var1, var2, listw,
zero.policy=NULL, nsim = 499,
alternative = "greater", scale = TRUE)
{
if(length(var1) != length(var2))
stop("calcSpatialCorrIndicators: var1 and var2 differ in length")
# if(length(var1) != length(hexlistw$neighbours))
# stop("calcSpatialCorrIndicators: length listw does not fit var1 and var2")
if(!(alternative %in% c("greater", "less", "two.sided")))
stop("calcSpatialCorrIndicators: alternative needs to be one of 'greater', 'less', 'two.side'")
# SSS for var 1
sssVar1 <- calcSSS(var1, listw = listw, zero.policy= zero.policy)
# SSS for var 2
sssVar2 <- calcSSS(var2, listw = listw, zero.policy= zero.policy)
# pearson's r for lagged var1 and lagged var2
rXlayLag <- calcPearsonLagged(var1, var2, listw = listw)
# Moran's I for var1
Ix <- moran.test(var1, listw = listw,
zero.policy= zero.policy,
alternative= alternative)$estimate[1] %>% as.numeric()
# Moran's I for var2
Iy <- moran.test(var2, listw = listw,
zero.policy= zero.policy,
alternative= alternative)$estimate[1] %>% as.numeric()
# Lee's L
leeVar1Var2 <- lee.mc(var1, var2, listw = listw,
zero.policy= zero.policy,
nsim = nsim,
alternative= alternative)$statistic %>% as.numeric()
# pearson's r for var1 and var2
rXY <- cor(var1, var2)
# bivariate Moran's I
bv_1_2 <- moran_bv(var1, var2, listw = listw,
nsim = nsim, scale = scale)$t0
bv_2_1 <- moran_bv(var2, var1, listw = listw,
nsim = nsim, scale = scale)$t0
res <- list(sssVar1 = sssVar1,
sssVar2 = sssVar2,
rXlayLag = rXlayLag,
Ix = Ix, Iy = Iy,
leeVar1Var2 = leeVar1Var2,
rXY = rXY,
bv_1_2 = bv_1_2,
bv_2_1 = bv_2_1)
return(res)
}printSpatialCorrIndicators <- function(indi, n.signif=3)
{
cat(paste("r =\t", indi$rXY %>% signif(n.signif)), "\n")
cat(paste("r_lx,ly =", indi$rXlayLag %>% signif(n.signif)), "\n")
cat(paste("SSS_x =\t", indi$sssVar1 %>% signif(n.signif)), "\n" )
cat(paste("SSS_y =\t", indi$sssVar2 %>% signif(n.signif)), "\n")
cat(paste("L_x,y =\t", indi$leeVar1Var2 %>% signif(n.signif)), "\n")
cat(paste("I_x =\t", indi$Ix %>% signif(n.signif)), "\n")
cat(paste("I_y =\t", indi$Iy %>% signif(n.signif)), "\n")
cat(paste("I_B(x,y) =", indi$bv_1_2 %>% signif(n.signif)), "\n")
cat(paste("I_B(y,x) =", indi$bv_2_1 %>% signif(n.signif)), "\n")
}8.6.1.2 Strong positive autocorrelation, same values
Here we are looking at the bivariate association of var1with itself.
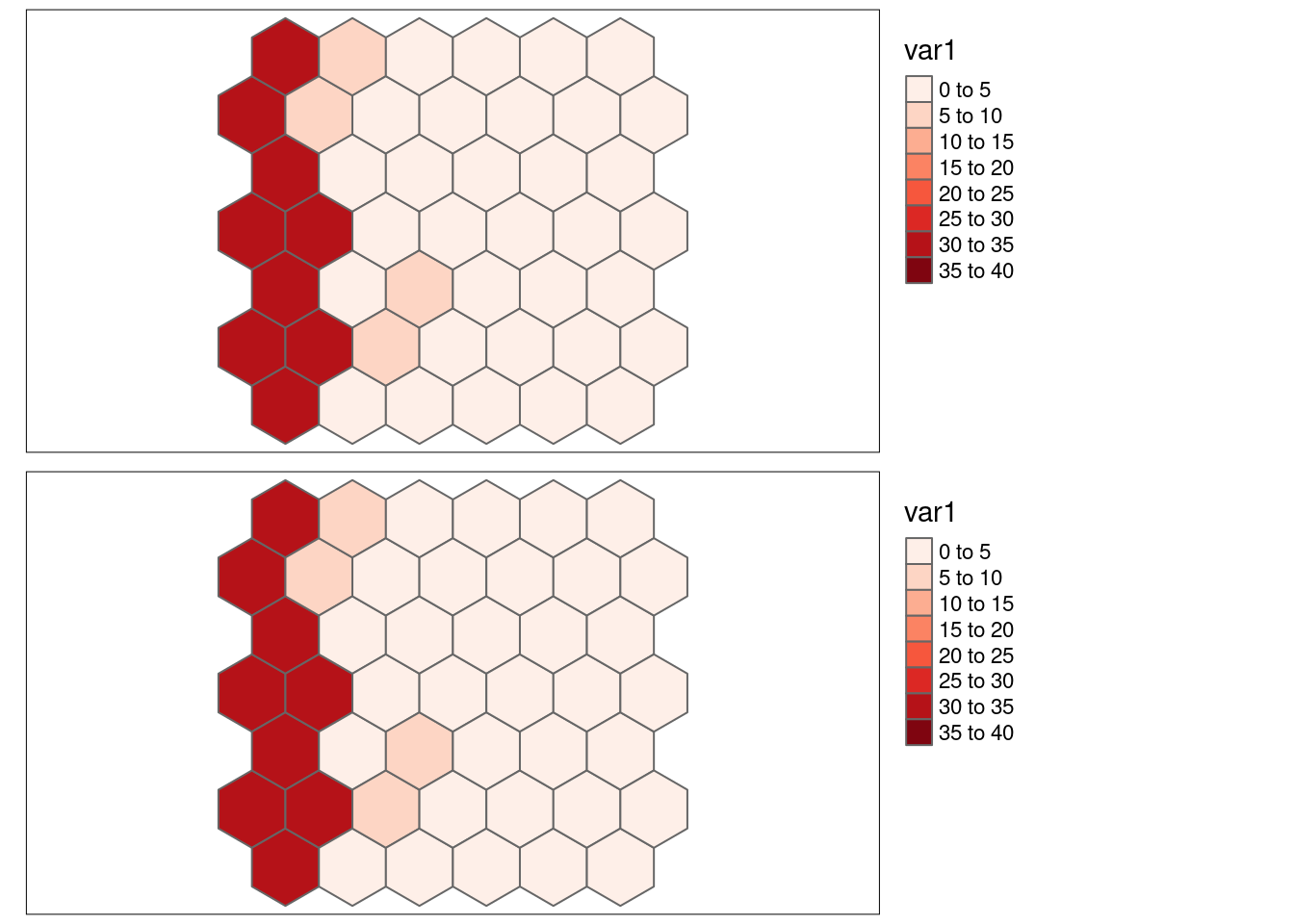
- \(SSS_{x}\) = 0.6112571
- \(SSS_{y}\) = 0.6112571
- \(r_{\tilde{x}, \tilde{y}}\) = 1
- \(L_{x,y}\) = 0.6112571
- \(I_x\) = 0.6778588
- \(I_y\) = 0.6778588
- r = 1
- \(L_{x,y}=\sqrt{SSS_x}\sqrt{SSS_y} \cdot r_{\tilde{x}, \tilde{y}}\)
- \(I_{B}(x,y)\) = 0.6778588
8.6.1.3 Strong positive autocorrelation, inverse values
var1neg has been created such that it shows a strong positive autocorrelation but the vaues are kind of inverted with respect to var1, leading to a negative Pearson’s r.
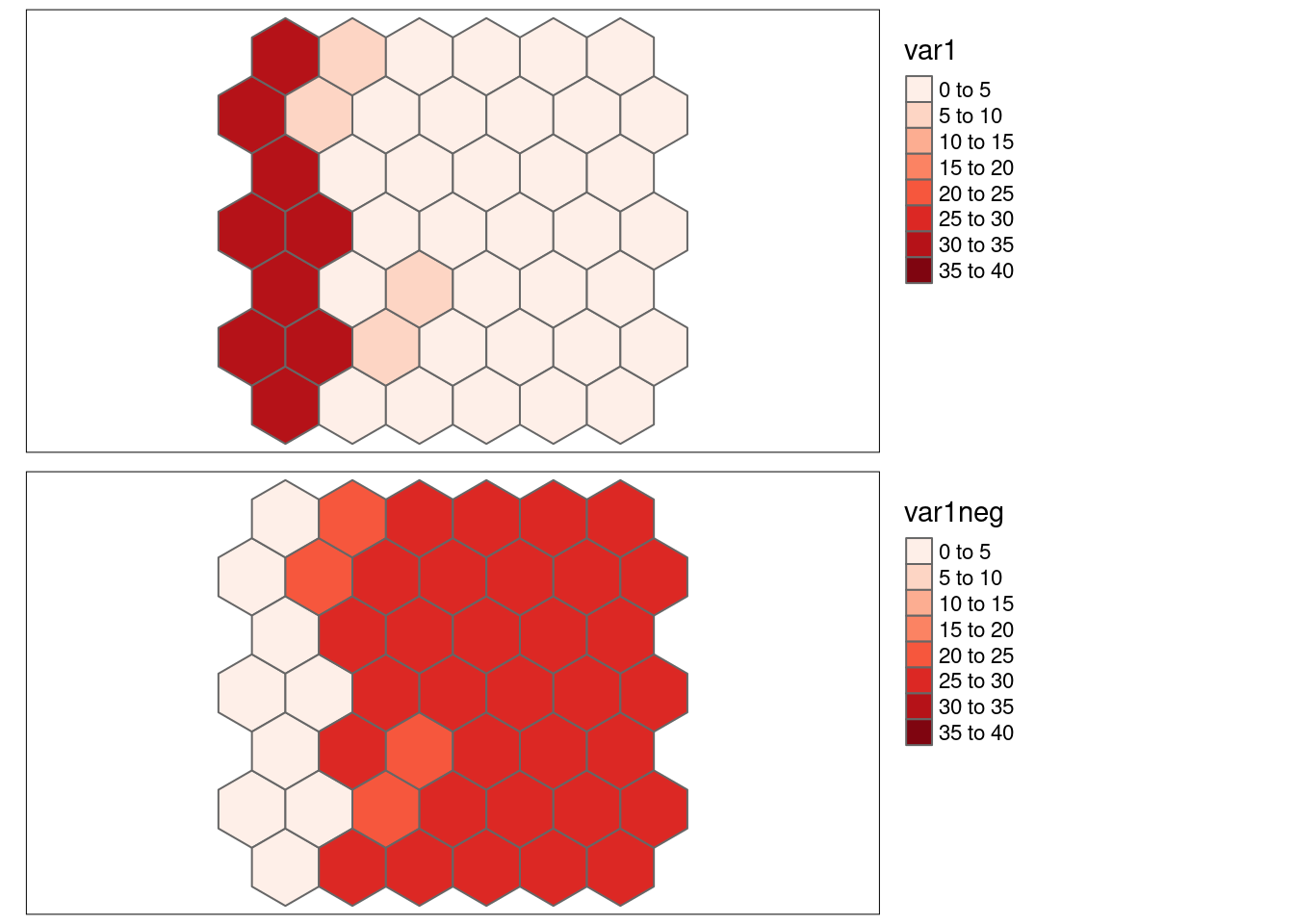
- \(SSS_{x}\) = 0.6112571
- \(SSS_{y}\) = 0.6112571
- \(r_{\tilde{x}, \tilde{y}}\) = -1
- \(L_{x,y}\) = -0.6112571
- \(I_x\) = 0.6778588
- \(I_y\) = 0.6778588
- r = -1
- \(I_{B}(x,y)\) = -0.6778588
8.6.1.4 Negative autocorrelation, same values
Here we look at the different measures for the bivariate association of an example with (weak) negative spatial autocorrelation var12 with it self.
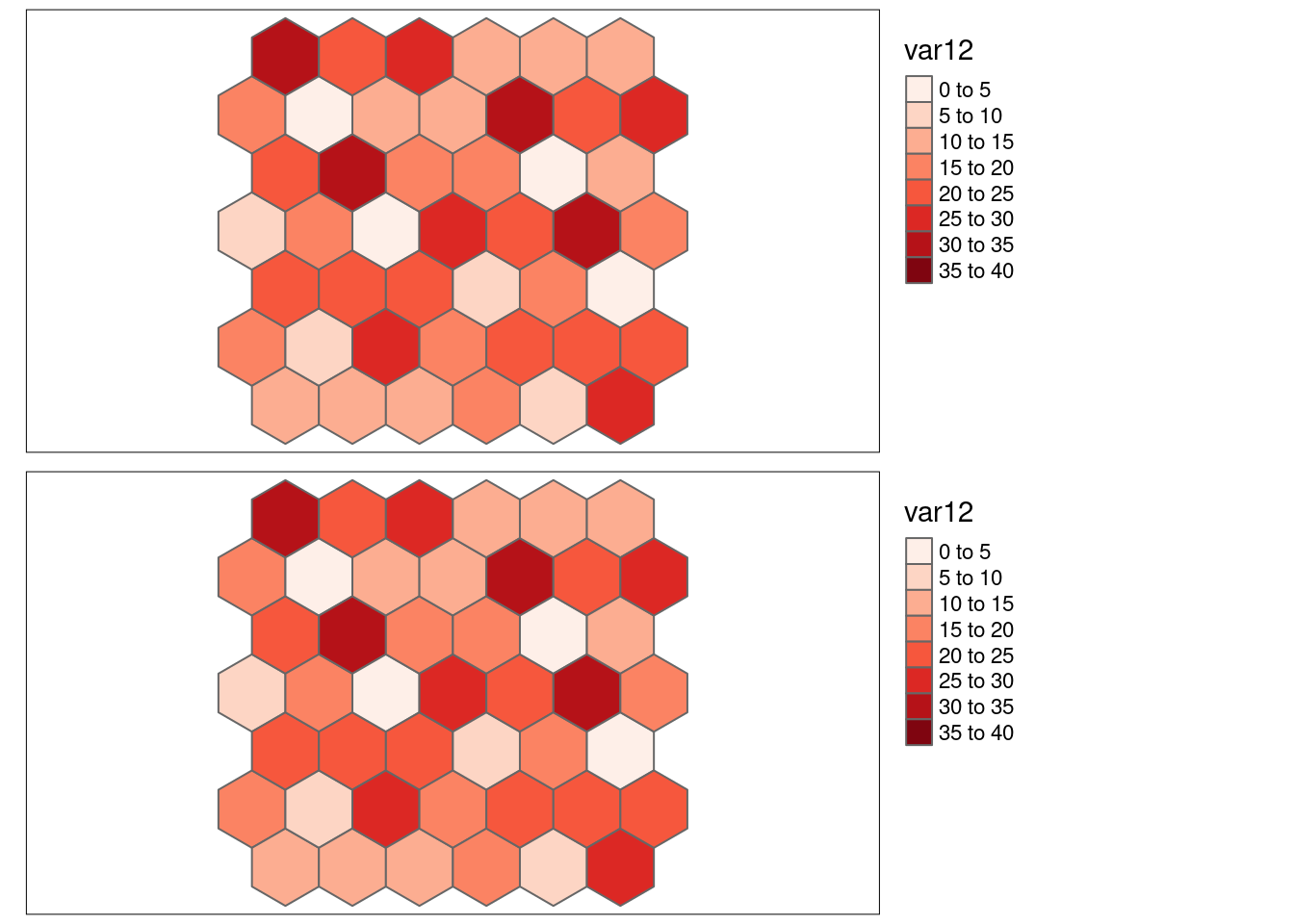
- \(SSS_{x}\) = 0.1340738
- \(SSS_{y}\) = 0.1340738
- \(r_{\tilde{x}, \tilde{y}}\) = 1
- \(L_{x,y}\) = 0.1340738
- \(I_x\) = -0.3083754
- \(I_y\) = -0.3083754
- r = 1
- \(I_{B}(x,y)\) = -0.3083754
8.6.1.5 Strong positive autocorrelation, contrary pattern
var2has been created such that it has a strong positive spatial autocorrelation but that high values of var2 are linked to low values for var1, leading to a negative association.
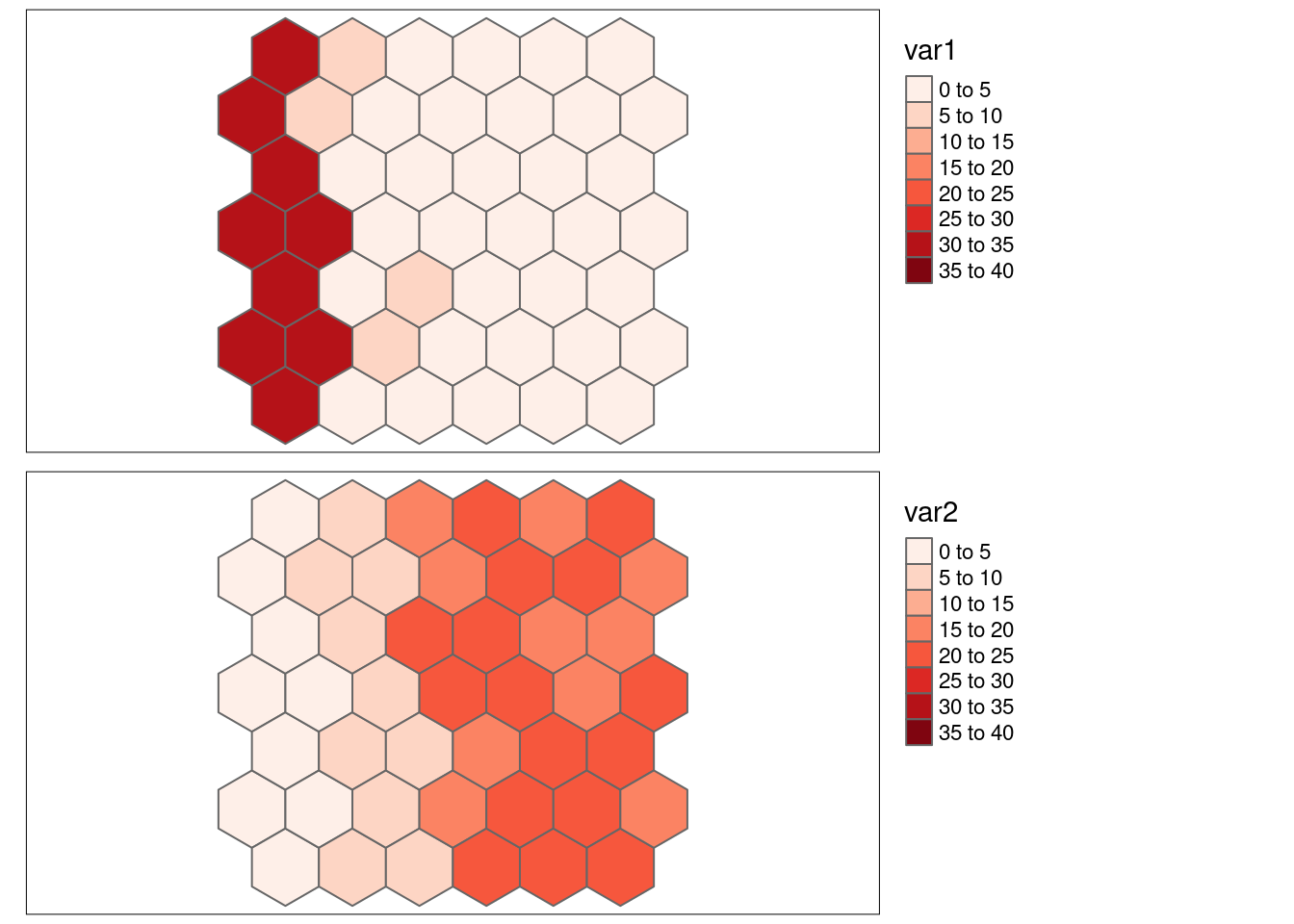
- \(SSS_{x}\) = 0.6112571
- \(SSS_{y}\) = 0.7654227
- \(r_{\tilde{x}, \tilde{y}}\) = -0.9281803
- \(L_{x,y}\) = -0.6346862
- \(I_x\) = 0.6778588
- \(I_y\) = 0.8140493
- r = -0.8017904
- \(I_{B}(x,y)\) = -0.6976358
- \(I_{B}(y,x)\) = -0.6725747
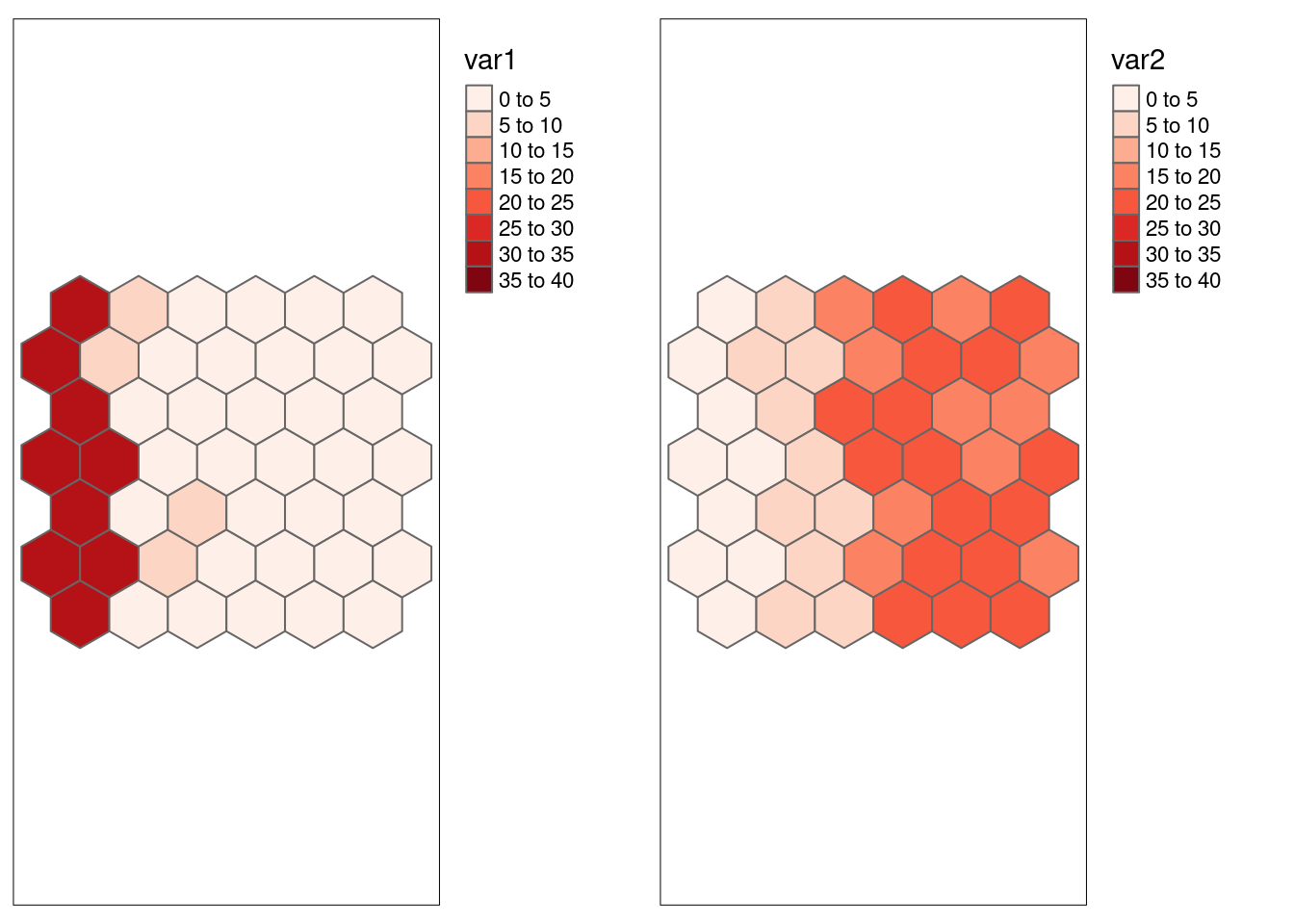
8.6.1.6 Example: var1 and var2, rearranged
Now we look at shuffled versions of var1 and var2: both variables were rearranged in exactly the same way, i.e. the observations still correspond. However, Moran’s I has changed as the shuffeling broke the spatial autocorrelation. var8 corresponds to the values in var1 and var9 to the values in var2.
The scatterplots look exactly the same:
p1 <- ggplot(hexgrid, aes(x=var1, y= var2)) +
geom_point() + labs(title = "Original")
p2 <- ggplot(hexgrid, aes(x=var8, y= var9)) +
geom_point() + labs(title = "Reshuffled")
ggarrange(p1,p2)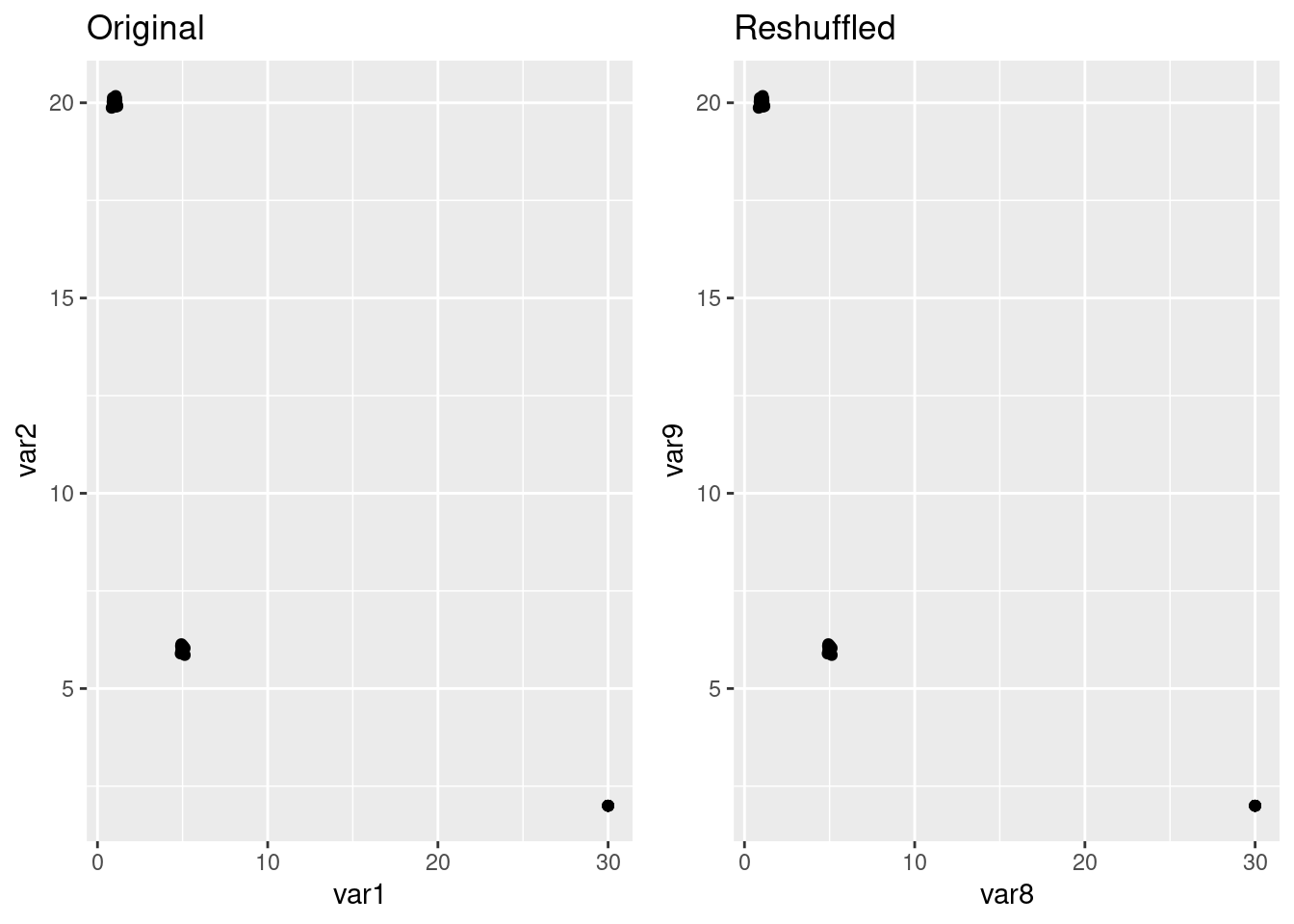
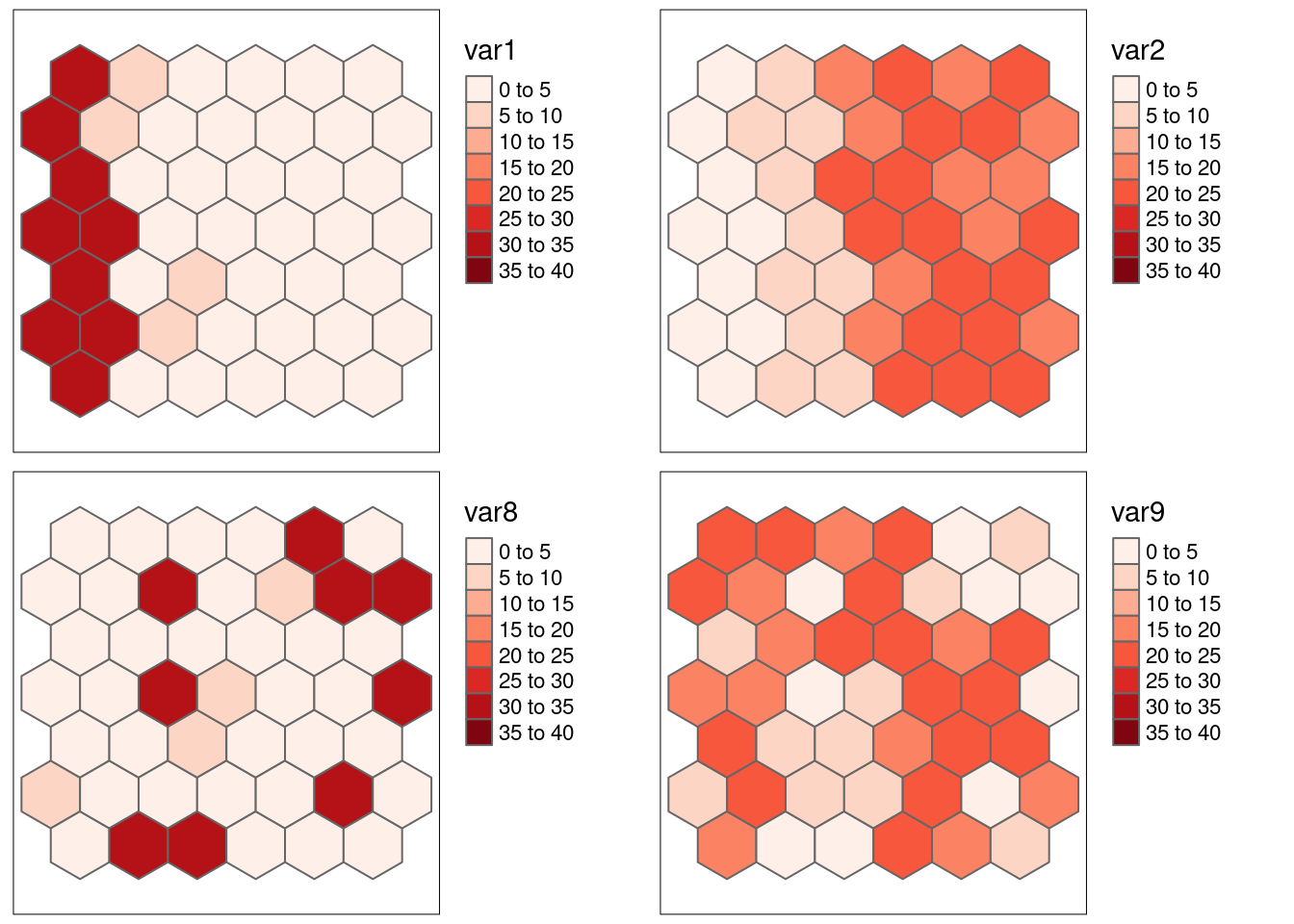
Let’s have a look at Moran’s I
varPos <- c(1,2,8,9)
moran4vars <- lapply(varPos+1, FUN = function(x) moran.test(st_drop_geometry(hexgrid)[,x], listw = hexlistw))
moran4varsShorted <- sapply(moran4vars, FUN = function(x) c(x$estimate[1], x$p.value))
row.names(moran4varsShorted)[2] <- "p-value"
colnames(moran4varsShorted) <- paste0("var", varPos)
t(moran4varsShorted)## Moran I statistic p-value
## var1 0.67785884 1.351183e-14
## var2 0.81404932 3.210743e-19
## var8 -0.03219664 5.409725e-01
## var9 0.05562954 2.027172e-01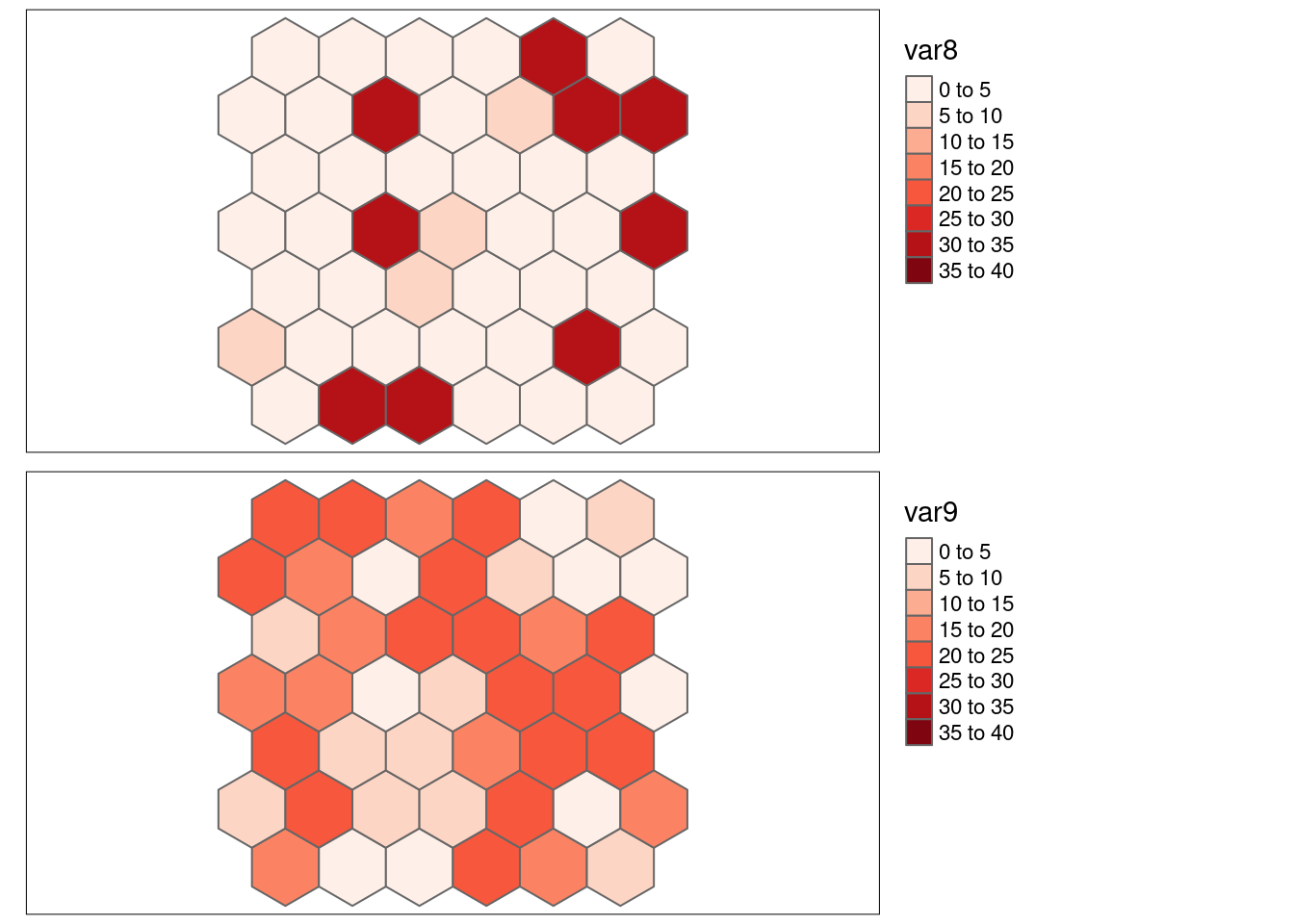 :::
::: {.column}
- \(SSS_{x}\) = 0.221982
- \(SSS_{y}\) = 0.2366627
- \(r_{\tilde{x}, \tilde{y}}\) = -0.8358157
- \(L_{x,y}\) = -0.1915687
- \(I_x\) = -0.0321966
- \(I_y\) = 0.0556295
- r = -0.8017904
- \(I_{B}(x,y)\) = -0.0200969
- \(I_{B}(y,x)\) = -0.0259268
:::
::: {.column}
- \(SSS_{x}\) = 0.221982
- \(SSS_{y}\) = 0.2366627
- \(r_{\tilde{x}, \tilde{y}}\) = -0.8358157
- \(L_{x,y}\) = -0.1915687
- \(I_x\) = -0.0321966
- \(I_y\) = 0.0556295
- r = -0.8017904
- \(I_{B}(x,y)\) = -0.0200969
- \(I_{B}(y,x)\) = -0.0259268
Lee’s L for var8 and var9 shows no significant spatial pattern of bivariate association.
##
## Monte-Carlo simulation of Lee's L
##
## data: hexgrid$var8 , hexgrid$var9
## weights: hexlistw
## number of simulations + 1: 500
##
## statistic = -0.19157, observed rank = 114, p-value = 0.228
## alternative hypothesis: lessLee’s L for var1 and var2 shows a clear negative bivariate spatial association. The positive spatial autocorrelation present in var1 and var2 increased Lee’ L.
##
## Monte-Carlo simulation of Lee's L
##
## data: hexgrid$var1 , hexgrid$var2
## weights: hexlistw
## number of simulations + 1: 500
##
## statistic = -0.63469, observed rank = 1, p-value = 0.002
## alternative hypothesis: lessPearson’s r did not change (by definition) between var1and var2and for var8and var9.
##
## Pearson's product-moment correlation
##
## data: hexgrid$var8 and hexgrid$var9
## t = -8.7979, df = 43, p-value = 3.624e-11
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## -0.8866491 -0.6646941
## sample estimates:
## cor
## -0.8017904The significance of the test for Pearson’s r is not really changed if the look at the modified t-test
var1 vs. var2
##
## Corrected Pearson's correlation for spatial autocorrelation
##
## data: x and y ; coordinates: X and Y
## F-statistic: 51.8097 on 1 and 28.7818 DF, p-value: 0
## alternative hypothesis: true autocorrelation is not equal to 0
## sample correlation: -0.8018var8 vs. var9
##
## Corrected Pearson's correlation for spatial autocorrelation
##
## data: x and y ; coordinates: X and Y
## F-statistic: 43.7545 on 1 and 24.3069 DF, p-value: 0
## alternative hypothesis: true autocorrelation is not equal to 0
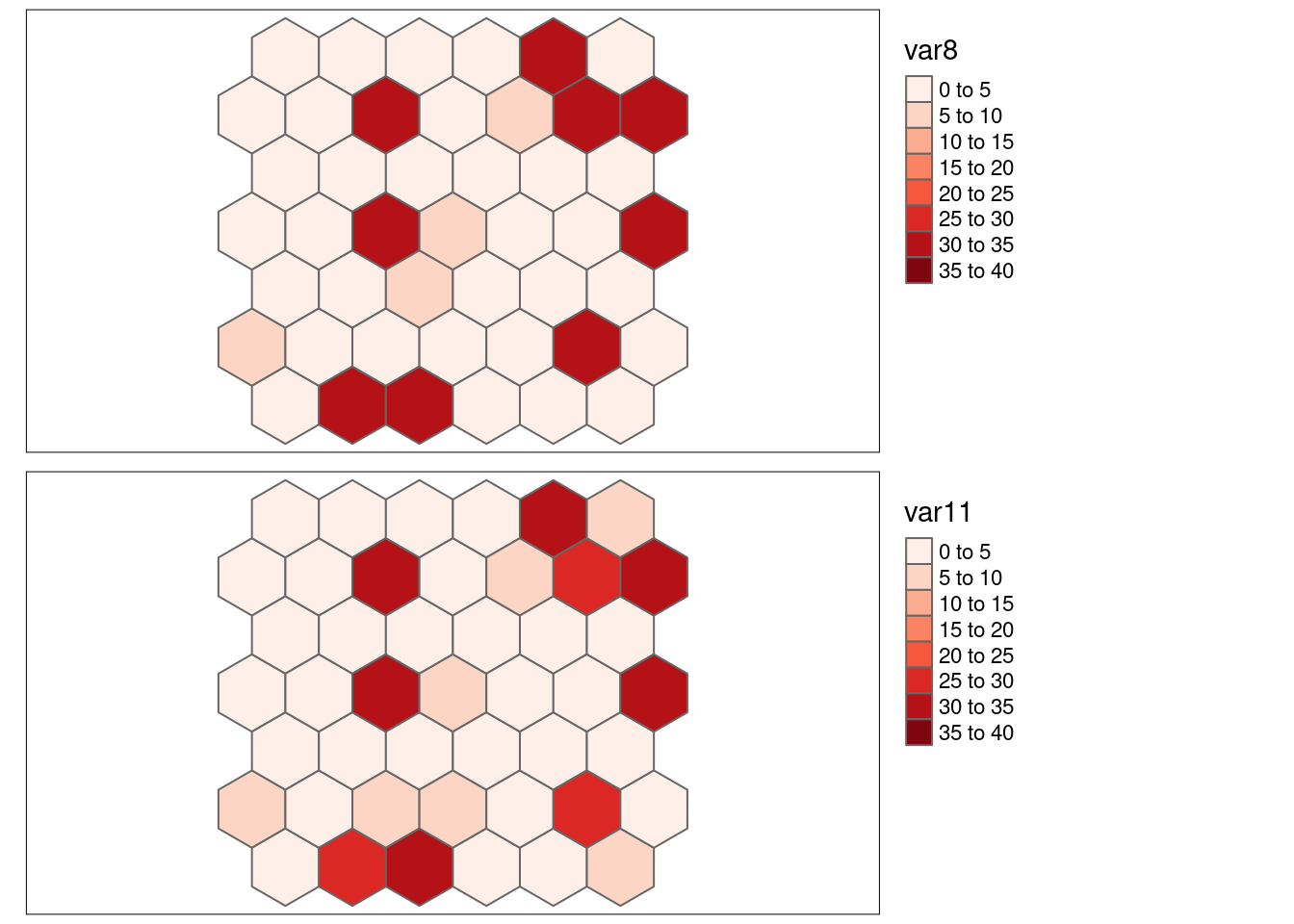
## sample correlation: -0.80188.6.1.7 Positive correlation with and without positive spatial autocorrelation var1-var10 and var8-var11 (reshuffled)
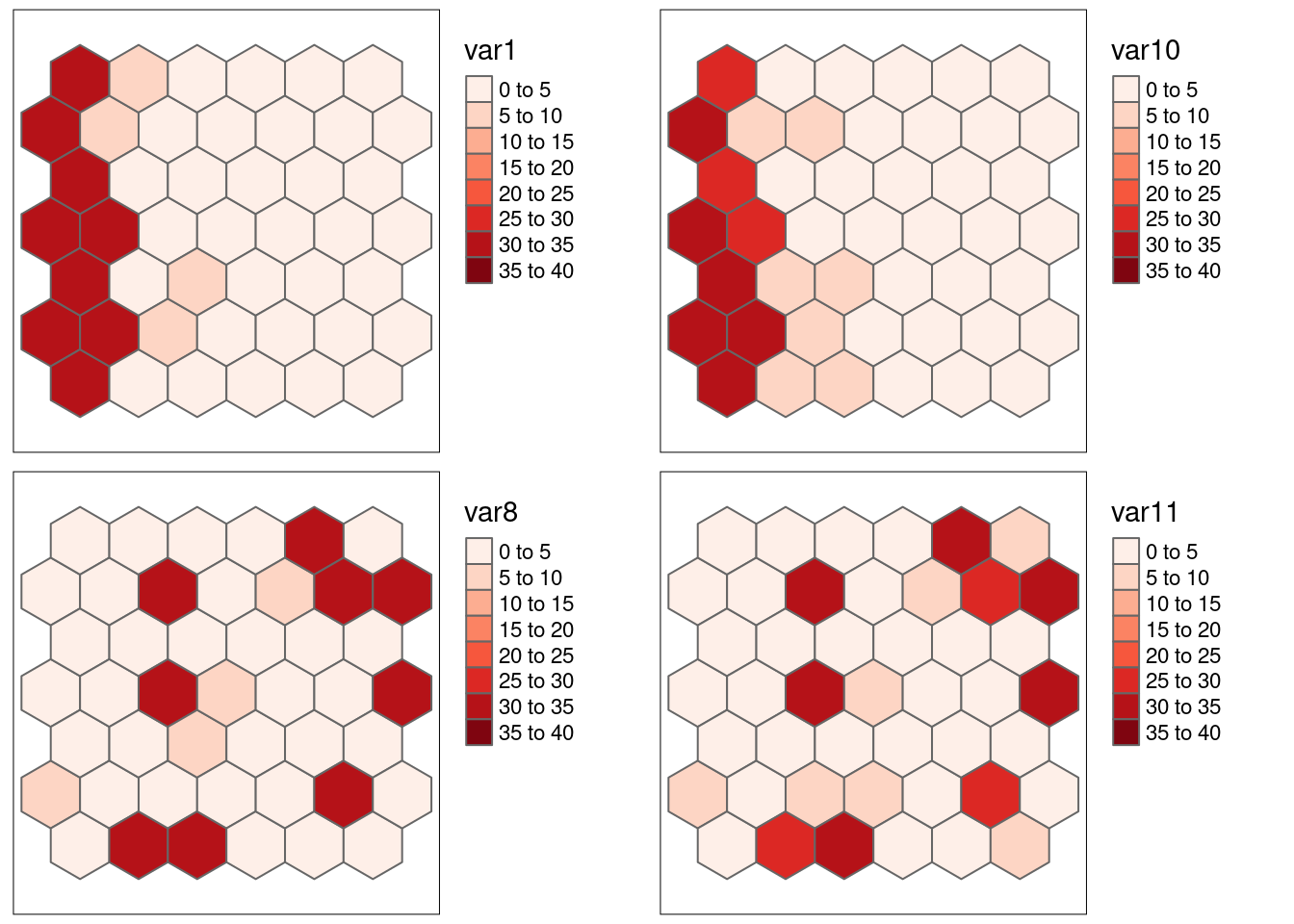
Scatterplot
p1 <- ggplot(hexgrid, aes(x=var1, y= var10)) +
geom_point() + labs(title = "Original")
p2 <- ggplot(hexgrid, aes(x=var8, y= var11)) +
geom_point() + labs(title = "Reshuffled")
ggarrange(p1,p2)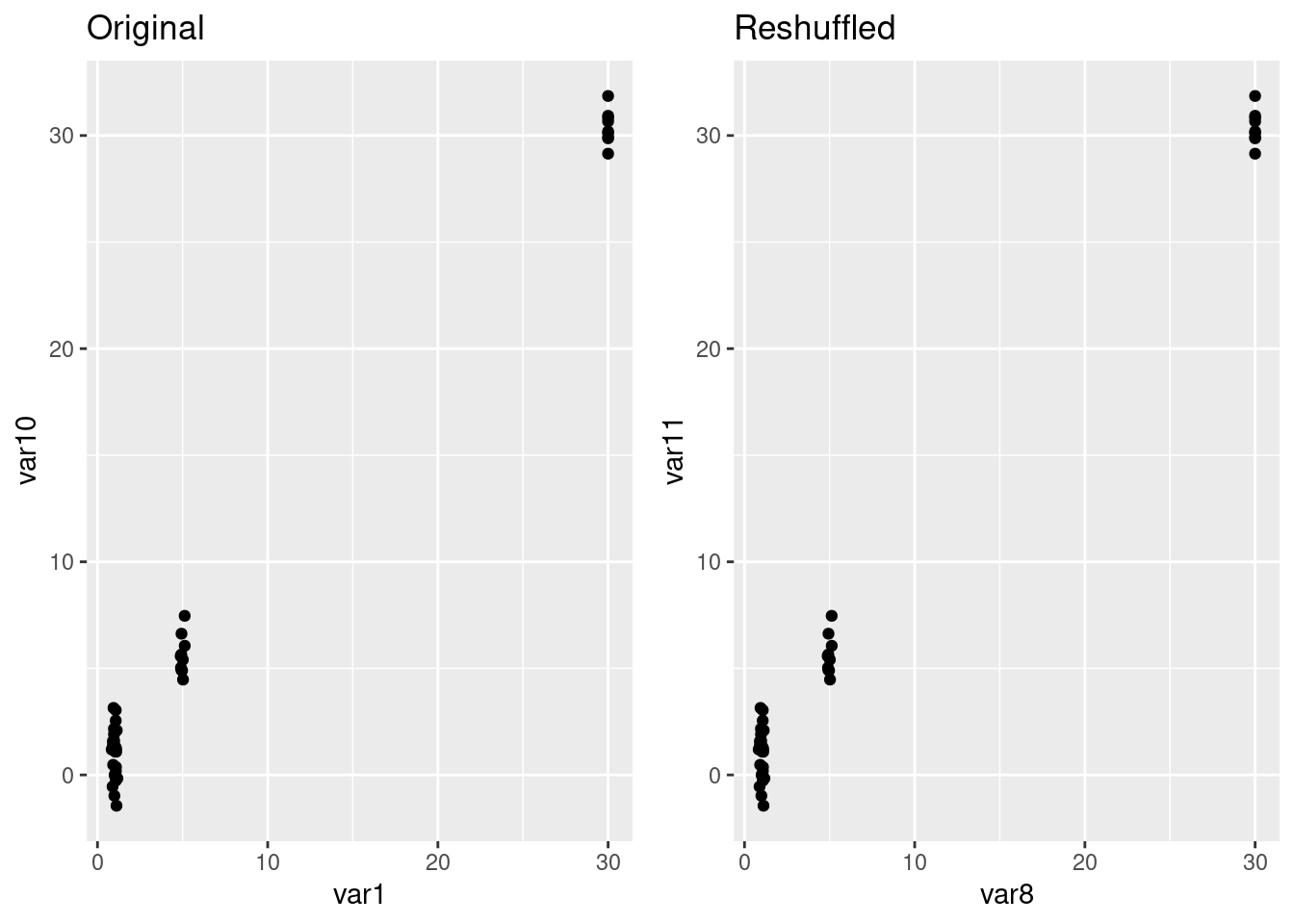
Pearson’s r var1 vs. var10
##
## Pearson's product-moment correlation
##
## data: hexgrid$var1 and hexgrid$var10
## t = 71.789, df = 43, p-value < 2.2e-16
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## 0.9924220 0.9977336
## sample estimates:
## cor
## 0.9958542Pearson’s r var8 vs. var11
##
## Pearson's product-moment correlation
##
## data: hexgrid$var8 and hexgrid$var11
## t = 71.789, df = 43, p-value < 2.2e-16
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## 0.9924220 0.9977336
## sample estimates:
## cor
## 0.9958542Lee’s L var1 vs. var10
##
## Monte-Carlo simulation of Lee's L
##
## data: hexgrid$var1 , hexgrid$var10
## weights: hexlistw
## number of simulations + 1: 500
##
## statistic = 0.61188, observed rank = 500, p-value < 2.2e-16
## alternative hypothesis: two.sided- spatial rearrangement makes the pattern less similar according to Lee’s L
- as the positive spatial autocorrelation in var1 and var10 is destroyed thereby
- Pearson’s r cannot capture that as it does not consider the spatial pattern
Lee’s L var8 vs. var11
##
## Monte-Carlo simulation of Lee's L
##
## data: hexgrid$var8 , hexgrid$var11
## weights: hexlistw
## number of simulations + 1: 500
##
## statistic = 0.21908, observed rank = 344, p-value = 0.624
## alternative hypothesis: two.sidedMoran’s I var1-var10 & var8-var11
varPos <- c(1,10,8,11)
moran4vars <- lapply(varPos+1, FUN = function(x) moran.test(st_drop_geometry(hexgrid)[,x], listw = hexlistw))
moran4varsShorted <- sapply(moran4vars, FUN = function(x) c(x$estimate[1], x$p.value))
row.names(moran4varsShorted)[2] <- "p-value"
colnames(moran4varsShorted) <- paste0("var", varPos)
t(moran4varsShorted)## Moran I statistic p-value
## var1 0.67785884 1.351183e-14
## var10 0.68308295 9.003126e-15
## var8 -0.03219664 5.409725e-01
## var11 -0.03448282 5.507866e-018.6.1.7.1 Strong positive autocorrelation, positive association
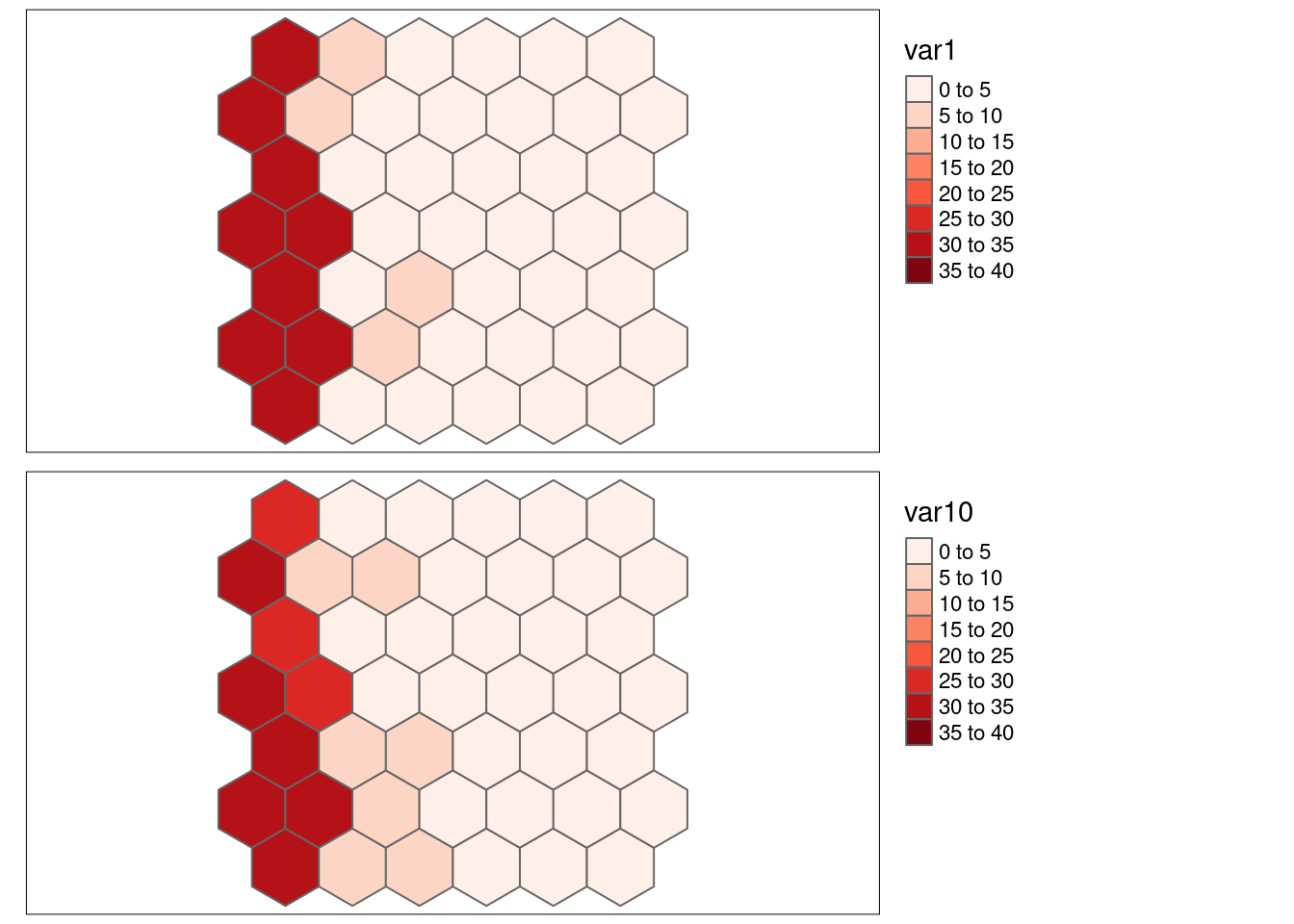
- \(SSS_{x}\) = 0.6112571
- \(SSS_{y}\) = 0.613609
- \(r_{\tilde{x}, \tilde{y}}\) = 0.9990976
- \(L_{x,y}\) = 0.6118786
- \(I_x\) = 0.6778588
- \(I_y\) = 0.6830829
- r = 0.9958542
- \(I_{B}(x,y)\) = 0.6794298
- \(I_{B}(y,x)\) = 0.6820743
8.6.1.8 Strong positive autocorrelation, shifted by one cell
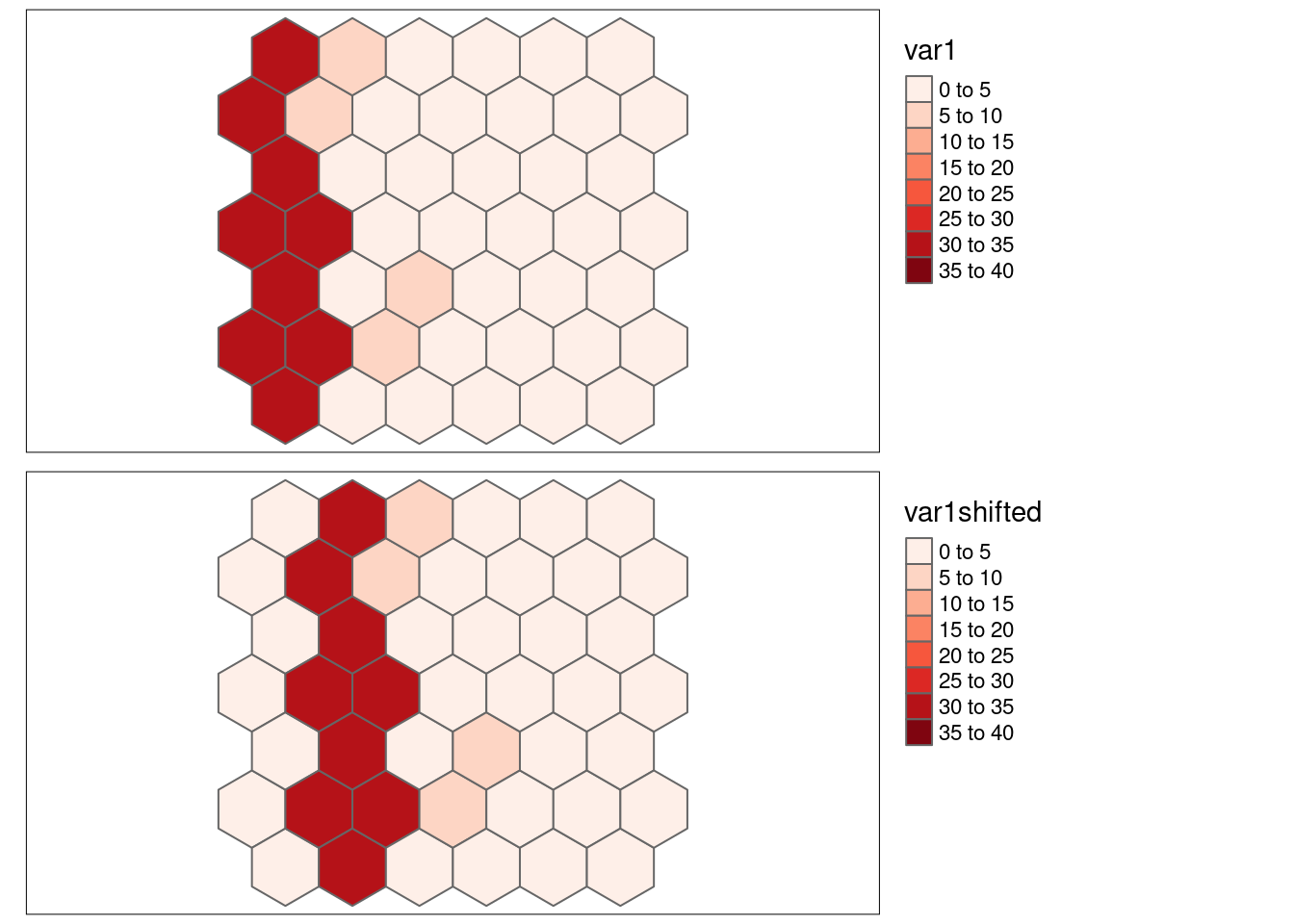
- \(SSS_{x}\) = 0.6112571
- \(SSS_{y}\) = 0.4151456
- \(r_{\tilde{x}, \tilde{y}}\) = 0.6660567
- \(L_{x,y}\) = 0.3317141
- \(I_x\) = 0.6778588
- \(I_y\) = 0.3445732
- r = 0.0919851
- \(I_{B}(x,y)\) = 0.3812871
- \(I_{B}(y,x)\) = 0.2708408
8.6.1.9 No autocorrelation, shifted by one cell
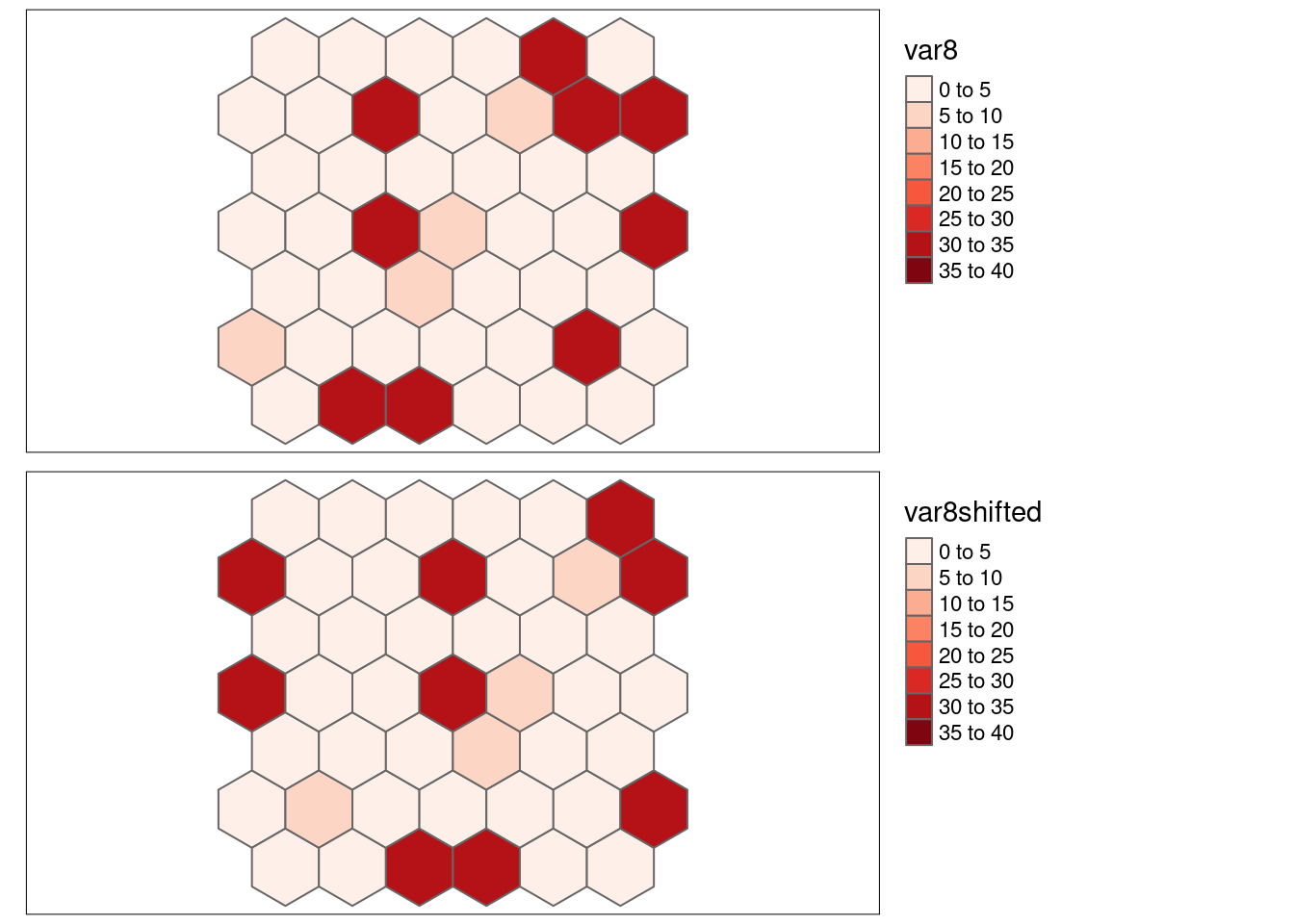
- \(SSS_{x}\) = 0.221982
- \(SSS_{y}\) = 0.1194423
- \(r_{\tilde{x}, \tilde{y}}\) = 0.2263546
- \(L_{x,y}\) = 0.0355491
- \(I_x\) = -0.0321966
- \(I_y\) = -0.0383465
- r = 0.0094375
- \(I_{B}(x,y)\) = 0.0742934
- \(I_{B}(y,x)\) = 0.1074713
8.7 The COVID-19 data case study - bivariate association
We will use the covid-19 incidence data for Germany that have been introduced previously.
## Reading layer `kreiseWithCovidMeldeWeeklyCleaned' from data source
## `/home/slautenbach/Documents/lehre/HD/git_areas/spatial_data_science/spatial-data-science/data/covidKreise.gpkg'
## using driver `GPKG'
## Simple feature collection with 400 features and 145 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: 280371.1 ymin: 5235856 xmax: 921292.4 ymax: 6101444
## Projected CRS: ETRS89 / UTM zone 32N## Warning: st_point_on_surface assumes attributes are constant over geometriesdistnb <- dnearneigh(poSurfKreise, d1=1, d2= 70*10^3)
unionedNb <- union.nb(distnb, polynb)
dlist <- nbdists(unionedNb, st_coordinates(poSurfKreise))
dlist <- lapply(dlist, function(x) 1/x)
unionedListw_d <- nb2listw(unionedNb, glist=dlist, style = "W")
ebiMc <- EBImoran.mc(n= st_drop_geometry(covidKreise)%>% pull("sumCasesTotal"),
x= covidKreise$EWZ, listw = unionedListw_d, nsim =999)## The default for subtract_mean_in_numerator set TRUE from February 20168.7.1 Incidence vs. votes for right winged party
Let’s look at the association between the transformed response variable (empirical bayes modification) with two variables we will be using later on as predictors in our model: the votes for the right winged party at the national election in 2017 (Stimmenanteile.AfD) and the rurality of a district (Laendlichkeitmeasured as the share of population that lives in densely populated areas in the district).
First the normal Pearson correlation coefficient:
##
## Pearson's product-moment correlation
##
## data: covidKreise$Stimmenanteile.AfD and ebiMc$z
## t = 10.226, df = 398, p-value < 2.2e-16
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## 0.3748636 0.5304747
## sample estimates:
## cor
## 0.4561491##
## Pearson's product-moment correlation
##
## data: covidKreise$Laendlichkeit and ebiMc$z
## t = 3.6908, df = 398, p-value = 0.0002547
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## 0.08538521 0.27505941
## sample estimates:
## cor
## 0.1819139When Lee’s L with the permutation test:
##
## Monte-Carlo simulation of Lee's L
##
## data: covidKreise$Stimmenanteile.AfD , ebiMc$z
## weights: unionedListw_d
## number of simulations + 1: 1000
##
## statistic = 0.35156, observed rank = 1000, p-value = 0.001
## alternative hypothesis: greater##
## Monte-Carlo simulation of Lee's L
##
## data: covidKreise$Laendlichkeit , ebiMc$z
## weights: unionedListw_d
## number of simulations + 1: 1000
##
## statistic = 0.11064, observed rank = 1000, p-value = 0.001
## alternative hypothesis: greaterWe see that the incorporation of spatial autocorrelation reduces the estimated strength of the relationship as both variables are positively spatially autocorrelated. As the spatial autocorrelation for rurality is weaker this leads to a lower difference bettwen Pearson’s r and Lee’s L.
##
## Monte-Carlo simulation of Moran I
##
## data: covidKreise$Stimmenanteile.AfD
## weights: unionedListw_d
## number of simulations + 1: 1000
##
## statistic = 0.73757, observed rank = 1000, p-value = 0.001
## alternative hypothesis: greater##
## Monte-Carlo simulation of Moran I
##
## data: covidKreise$Laendlichkeit
## weights: unionedListw_d
## number of simulations + 1: 1000
##
## statistic = 0.22267, observed rank = 1000, p-value = 0.001
## alternative hypothesis: greaterIn summary:
both variables (weakly) positively correlated
spatial lag of both variables also (weakly) positively correlated
both variables strong positive global spatial autocorrelation
positive bivariate spatial dependence (similarity)
r = -0.364
\(r_{\tilde{x}, \tilde{y}}\) = -0.657
\(I_x\) = -0.345
\(I_y\) = -0.345
\(SSS_{x}\) = 0.212
\(SSS_{y}\) = 0.212
\(L_{x,y}\) = -0.138
\(I_{B}(x,y)\) = 0.155
\(I_{B}(y,x)\) = 0.155
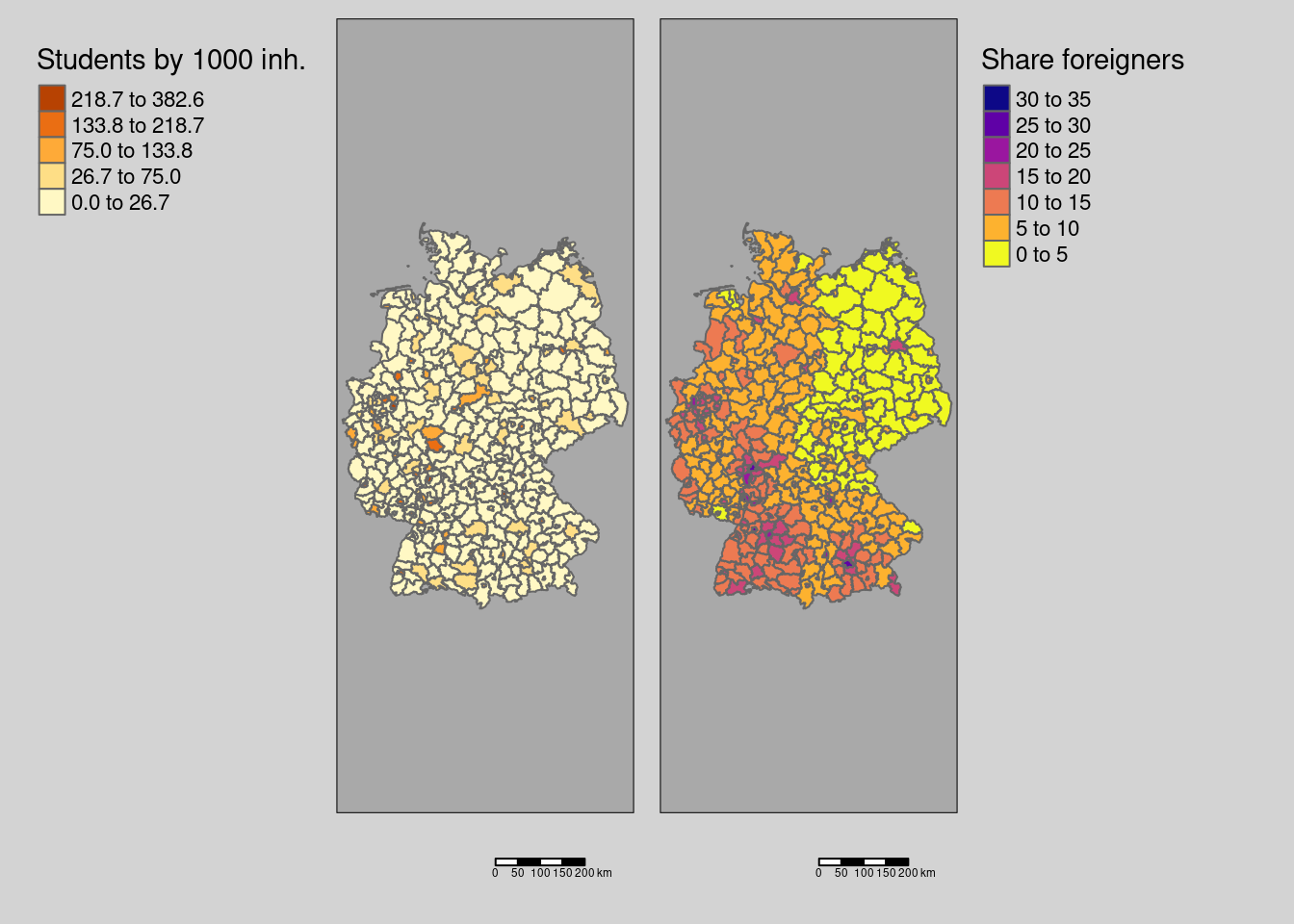
8.7.2 Fast internet availibility and rurality
covm1 <- tm_shape(covidKreise) + tm_polygons(fill="Breitbandversorgung",
fill.legend = tm_legend(title = "Fast internet", reverse=TRUE)) +
tm_layout(legend.outside = TRUE, bg.color = "darkgrey", outer.bg.color = "lightgrey",
legend.outside.size = 0.5, attr.outside = TRUE,
legend.outside.position = "left") + tm_scalebar()
covm2 <- tm_shape(covidKreise) + tm_polygons(fill="Laendlichkeit",
fill.scale = tm_scale_intervals(values= "-matplotlib.plasma"),
fill.legend = tm_legend(title = "Rurality", reverse=TRUE)) +
tm_layout(legend.outside = TRUE, bg.color = "darkgrey", outer.bg.color = "lightgrey",
legend.outside.size = 0.5, attr.outside = TRUE,
legend.outside.position = "right") + tm_scalebar()
tmap_arrange(covm1, covm2)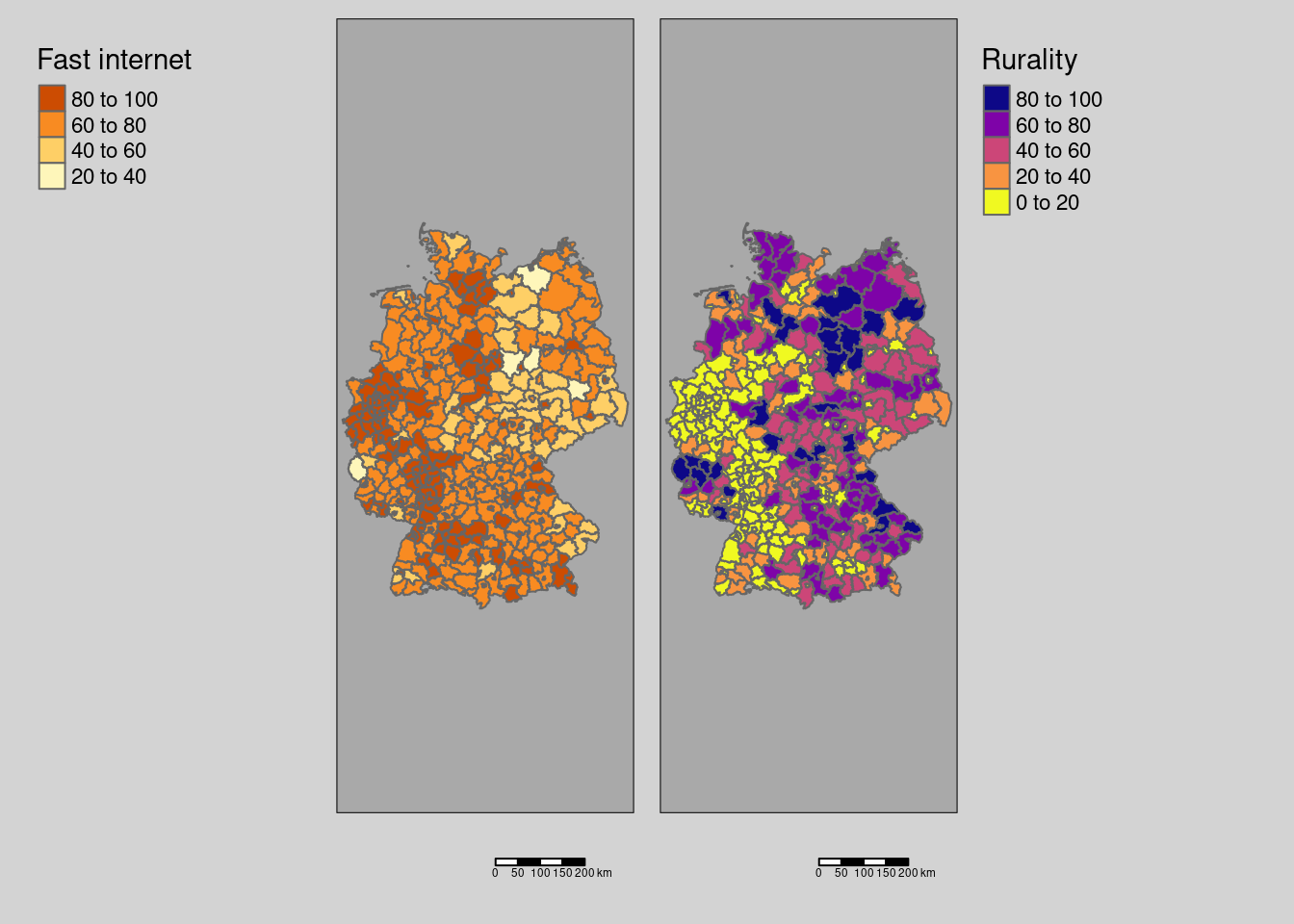
indi <- calcSpatialCorrIndicators(covidKreise$Breitbandversorgung, covidKreise$Laendlichkeit, listw = unionedListw_d)both variables negatively correlated
spatial lag of both variables also negatively correlated
both variables weak positive global spatial autocorrelation
negative bivariate spatial dependence (disimilarity)
\(r_{x,y}\) = -0.741
\(r_{\tilde{x}, \tilde{y}}\) = -0.783
\(I_x\) = 0.233
\(I_y\) = 0.223
\(SSS_{x}\) = 0.303
\(SSS_{y}\) = 0.267
\(L_{x,y}\) = -0.223
\(I_{B}(x,y)\) = -0.168
\(I_{B}(y,x)\) = -0.166